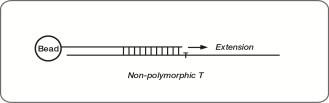
Specificity Controls
These controls are designed to monitor potential nonspecific primer extension for Infinium I and Infinium II assay probes. Specificity controls are designed against nonpolymorphic T sites.
These controls are designed to monitor allele-specific extension for Infinium I probes. The methylation status of a particular cytosine is carried out following bisulfite treatment of DNA by using query probes for unmethylated and methylated state of each CpG locus. In assay oligo design, the A/T match corresponds to the unmethylated status of the interrogated C, and the G/C match corresponds to the methylated status of C. G/T mismatch controls check for nonspecific detection of methylation signal over unmethylated background. PM controls correspond to A/T perfect match and should give high signal. MM controls correspond to G/T mismatch and should give low signal. Performance of GT mismatch controls should be monitored in both green and red channels. The Controls dashboard table lists expected outcomes for control probes.
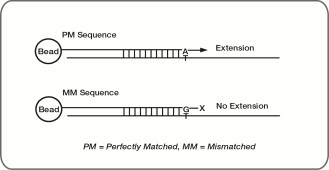
These controls are designed to monitor extension specificity for Infinium II probes and check for potential nonspecific detection of methylation signal over unmethylated background. Specificity II probes should incorporate the A base across the nonpolymorphic T and have intensity in the red channel. If there is nonspecific incorporation of the G base, the probe has elevated signal in the green channel.