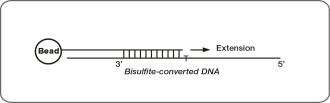
Bisulfite-Conversion Controls
These controls assess the efficiency of bisulfite conversion of the genomic DNA. The Infinium Methylation probes query a [C/T] polymorphism created by bisulfite conversion of non-CpG cytosines in the genome.
These controls use the Infinium I probe design and allele-specific single base extension to monitor efficiency of bisulfite conversion. If the bisulfite conversion reaction was successful, the C (converted) probes match the converted sequence and are extended. If the sample has unconverted DNA, the U (unconverted) probes are extended. There are no underlying C bases in the primer landing sites, except for the query site itself. Performance of bisulfite conversion controls C1, C2, and C3 should be monitored in the green channel. Controls C4, C5, and C6 should be monitored in red channel.
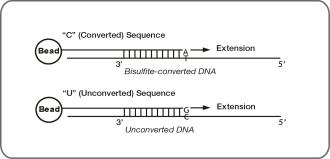
These controls use Infinium II probe design and single base extension to monitor the efficiency of bisulfite conversion. If the bisulfite conversion reaction was successful, the A base is incorporated and the probe has intensity in the red channel. If the sample has unconverted DNA, the G base is incorporated across the unconverted cytosine, and the probe has elevated signal in the green channel.