System Overview
This section provides an overview of the MiSeq i100 Plus System, including information on hardware, software, data analysis, and run management. For detailed specifications, data sheets, applications, and related products, refer to the MiSeq i100 Plus System support site.
|
Feature |
Description |
|---|---|
|
XLEAP SBS Chemistry |
The MiSeq i100 Plus System uses XLEAP SBS chemistry, which produces high quality data with fast sequencing run times compared to standard SBS run times. These performance improvements are achieved through an improved nucleotide blocker/linker, and a higher fidelity faster polymerase for nucleotide incorporation. |
|
Patterned Flow Cell |
The MiSeq i100 Plus System uses patterned flow cells, which are designed to enhance sequencing quality and efficiency. Patterned flow cells are compromised of nanowells containing complementary DNA probes at fixed specific locations on the surface of the flow cell. This feature eliminates the need to map cluster sites, speeds up sequencing time, and optimizes the use of available space on the flow cell. Due to the way the percentage of clusters passing filter (%PF) is calculated, instruments with patterned flow cells display lower %PF values compared to non-patterned flow cells. Despite the lower %PF, the overall yield is not impacted. |
|
CMOS |
The MiSeq i100 Plus System uses a patterned flow cell with nanowells integrated onto a CMOS chip. Each nanowell is aligned with a photodiode that detects light emissions at the bottom of the well, allowing for faster sequencing turnaround time. |
|
2-Channel |
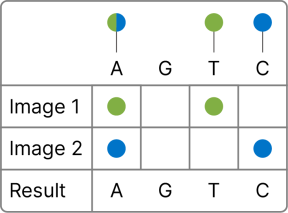
The MiSeq i100 Plus System uses two color chemistry, enabling rapid imaging of the flow cell using blue and green channels at each sequencing cycle. A feature of the MiSeq i100 Plus System is the excitation/emission strategy, which uses 2-channel excitation and 1-channel emission, further accelerating sequencing turnaround times.
A—Clusters with signals in green and blue. G—Clusters without signal in green or blue. T—Clusters with signal in green only. C—Clusters with signal in blue only. |
|
Index-First Sequencing |
The MiSeq i100 Plus System uses Index-first sequencing, allowing users to evaluate demultiplexing data within three hours from the start of a run. Index-first sequencing allows for same-day adjustments to be made for subsequent run planning if necessary. |
|
Room temperature consumables |
The MiSeq i100 Plus System consumables are shipped and stored at ambient temperatures, resulting in reduced packaging, easy consumable preparation, and eliminate the need for cold storage units. |
|
On board denaturation |
The MiSeq i100 Plus System accommodates single-stranded and double-stranded templates for sequencing. Template library preparation involves dilution with buffers, provided in each sequencing kit, which are loaded onto the sequencing consumable. The template is denatured onboard, which reduces workflow complexity. |
|
Illumina Run Manager |
Illumina Run Manager is integrated in the MiSeq i100 Plus Control Software, which enables run planning, run review, and management of select settings remotely using a web browser. Refer to Illumina Run Manager. |
|
Kiosk-mode |
MiSeq i100 Plus System features a Kiosk mode to enhance the security of the system to prevent unauthorized users to access the operating system. If an Administrator has to access the operating system to install a third-party application, such as a virus scanner, contact Illumina to get a temporary access code to access the operating system. |
|
DRAGEN compression |
DRAGEN ORA Compression is a fully lossless compression with a higher compression ratio than *.fastq.gz. Refer to the DRAGEN ORA support site. |
|
Feature |
Description |
|---|---|
|
Library Quality |
Adapter/primer dimers, partial library constructs, and contaminants can compromise the data quality and sequencing yield. Capillary electrophoresis methods (for example, Bioanalyzer, Fragment Analyzer, or Tape station) can be used for quality control and to visualize undesired library preparation remnants. An additional bead purification step can be used to remove the contaminants. |
|
Library Quantification |
Accurate library quantification is essential for optimal template loading onto the system. For best results, adhere to the recommendations for quantification provided in the library preparation guide. If guidance is not provided, use quantifying libraries by size-normalized qPCR for consistency and accuracy. |
|
Loading concentration |
Perform titration runs to identify the optimal loading concentration. When optimizing loading concentration, center titration experiments at 100 pM and fine tuning in 25-50 pM increments. |
|
Nucleotide Diversity |
Libraries with low nucleotide diversity can negatively impact template registration, data quality, and yield. To compensate for low base diversity in libraries, spike in the PhiX Control v3 Library sequencing. Titration experiments may be needed to identify the quantity of spike in required for optimal performance. |
|
Insert Size Representation |
For some libraries, insert size can decrease as the loading concentration increases. The optimal range for your library and application can vary depending on your workflow requirements. |