Pool and Denature Libraries
Sequencing System: NovaSeq X Plus
Library Prep: Illumina Cell-Free DNA Prep with Enrichment, manual normalization
Protocol L
Use this protocol to denature and dilute Illumina Cell-Free DNA Prep with Enrichment libraries prepared using the manual normalization option.
The NovaSeq X Plus 10B Flow Cell has eight lanes and the NovaSeq X Plus 1.5B Flow Cell has two lanes. Use this protocol for sequencing a single pool across both lanes of the 1.5B Flow Cell or a single lane of the 10B Flow Cell.
This protocol requires the following consumables and equipment.
Consumables
|
Consumables |
Supplier |
Purpose |
|---|---|---|
|
Low DNA binding microcentrifuge tube, 1.5 ml |
General lab supplier |
Combining volumes when pooling |
|
Microcentrifuge tube, 1.5 ml |
VWR, catalog # 20170-038, or equivalent |
Combining volumes |
|
Pipette tips, 20 μl |
General lab supplier |
Pipetting reagents and libraries. |
|
Pipette tips, 200 μl |
General lab supplier |
Pipetting reagents and libraries. |
|
Pipette tips, 1000 μl |
General lab supplier |
Pipetting reagents and libraries. |
|
Pre-load buffer |
Illumina, provided in the NovaSeq X Series reagent kit |
Neutralizing denatured libraries. |
|
Resuspension Buffer (RSB) |
Illumina, provided in the library prep kit contents |
Diluting libraries. |
Equipment
|
Equipment |
Supplier |
||||||
|---|---|---|---|---|---|---|---|
|
Heat block for 1.5 ml microcentrifuge tubes. The heat block must meet the following performance specifications:
|
General lab supplier |
||||||
|
Microcentrifuge |
General lab supplier |
||||||
|
Vortexer |
General lab supplier |
Prepare NaOH
Prepare a fresh dilution of 0.2 N NaOH to denature libraries for sequencing. Extra volume is prepared to prevent small pipetting errors from affecting the final NaOH concentration.
| 1. | [10B] Combine the following volumes in a microcentrifuge tube. |
|
Reagent |
Volume for One 10B Flow Cell (µl) |
Volume for Two 10B Flow Cells (µl) |
|---|---|---|
|
Laboratory-grade water |
80 |
160 |
|
Stock 1 N NaOH |
20 |
40 |
These volumes result in 100 µl 0.2 N NaOH for one flow cell or 200 µl 0.2 N NaOH for two flow cells.
| 2. | [1.5B] Combine the following volumes in a microcentrifuge tube. |
|
Reagent |
Volume for One 1.5B Flow Cell (µl) |
Volume for Two 1.5B Flow Cells (µl) |
|---|---|---|
|
Laboratory-grade water |
20 |
40 |
|
Stock 1 N NaOH |
5 |
10 |
These volumes result in 25 µl 0.2 N NaOH for one flow cell or 50 µl 0.2 N NaOH for two flow cells.
| 3. | Vortex, and then centrifuge the tube to mix. |
Normalize Libraries
Perform the following procedure to dilute libraries to the starting concentration. It is recommended to dilute only the amount that is needed to perform sequencing.
| 1. | Calculate the molarity value of the library using the following formula. |
| • | It is recommended to use a fluorometric quantification method that uses DNA binding dyes such as the Qubit Fluorometer 4.0. |
| • | Use 350 bp as the average library size. |
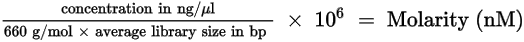
| 2. | Using the molarity value, calculate the volume of RSB and library needed to dilute each library to a starting concentration of 10 nM. |
| 3. | Dilute each library to the starting concentration using RSB. |
Pool Normalized Libraries
| 1. | Calculate library plexity for sequencing. Select the appropriate plexity calculation factor. |
|
Flow Cell Type |
Plexity Calculation Factor |
|---|---|
|
1.5B |
9.2 |
|
10B (single lane) |
6.8 |
| 2. | Divide the plexity calculation factor by the panel size for enrichment in megabases (Mb). |
This calculation is a recommended guideline. If each sequenced library does not achieve at least 30,000x on target coverage, reduce the number of libraries in the pool.
The maximum number of libraries that can be sequenced together is 192, due to limitation of individual indexes.
| 3. | Gently pipette to mix each normalized library 5 times. |
Use fresh tips for each library.
| 4. | Label a 1.5 ml low DNA binding microcentrifuge tube PDL (pooled DNA libraries). |
| 5. | Transfer equal volumes of each normalized library to the PDL tube. The recommended final pool volume is a minimum of 25 μl. For example, for a 10-plex library pool, combine 2.5 µl from each normalized library. |
A library generated as a 4-plex enrichment pool is 4x less concentrated than a 1-plex library. Therefore, sequencing pooling by volume should respect that ratio. For example, 20 μl of 4-plex libraries should be pooled with 5 μl of a 1-plex library.
| 6. | Vortex the PDL tube briefly to mix, and then centrifuge briefly. |
Dilute Library Pool
| 1. | Label a 1.5 ml low DNA binding microcentrifuge tube DIL1 (Dilution 1). |
| 2. | Label a 1.5 ml low DNA binding microcentrifuge tube DIL2 (Dilution 2). |
| 3. | Combine the following volumes in the DIL1 tube: |
| • | PDL (7.5 μl) |
| • | RSB (142.5 μl) |
These volumes produce a 150 μl library pool at 500 pM concentration.
| 4. | Vortex the DIL1 tube briefly to mix, and then centrifuge briefly. |
| 5. | Combine the following volumes in the DIL2 tube: |
| • | DIL1 (45 μl) |
| • | RSB (105 μl) |
These volumes produce a 150 μl library pool at 150 pM concentration.
| 6. | Vortex the DIL2 tube briefly to mix, and then centrifuge briefly. |
Denature Libraries
| 1. | Label a new 1.5 ml low DNA binding microcentrifuge tube LOAD. |
| 2. | Add the appropriate volume of DIL2 and 0.2 N NaOH to the LOAD tube as follows. |
|
Flow Cell Type |
DIL2 (µl) |
0.2 N NaOH (µl) |
Resulting Volume (µl) |
|---|---|---|---|
|
1.5B |
74.8 |
18.7 |
93.5 |
|
10B (single lane) |
34 |
8.5 |
42.5 |
| 3. | Cap and then vortex briefly. |
| 4. | Centrifuge at 280 × g for up to 1 minute. |
| 5. | Incubate at room temperature for 5 minutes to denature. |
| 6. | Add the appropriate volume of Pre-load buffer as follows. |
|
Flow Cell Type |
Pre-load Buffer (µl) |
Resulting Volume (µl) |
|---|---|---|
|
1.5B |
280.5 |
374 |
|
10B (single lane) |
127.5 |
170 |
| 7. | Cap and then vortex briefly. |
| 8. | Centrifuge at 280 × g for up to 1 minute. |
| 9. | Store libraries on ice until transferred to the library tube strip. |
After denaturing and diluting the libraries, you are ready to load the lyo insert and library tube strip into the reagent cartridge in preparation for sequencing. Refer to the sequencing system product documentation for more information.
Revision History - Protocol for Illumina Cell-Free DNA Prep with Enrichment, Manual Normalization
|
Document |
Date |
Description of Change |
|---|---|---|
|
Document # 200055977 v00 |
June 2024 |
Initial release |
