Introduction
The Illumina Single Cell 3′ RNA Prep, T100 kit in combination with the Illumina Single Cell Supplemental Enrichment and Amplification (SEA) Kit facilitates custom single cell RNA-Seq applications at the
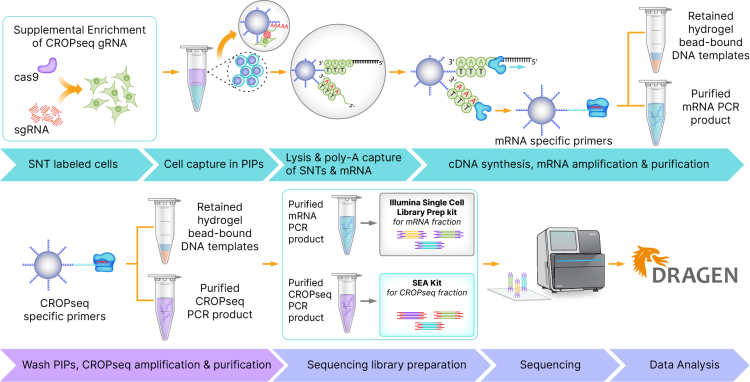
A common application of single-cell RNA sequencing is the capture of synthetic polyadenylated DNA tags (SNTs) that have been introduced to label select targets in a sample. The synthetic tags used and the methods for labeling unique samples with synthetic tags depends on user needs. The SEA Kit enables the amplification and isolation of SNTs such as pooled single guide RNA (sgRNA) products to generate sequencing-ready libraries compatible with Illumina Single Cell 3′ RNA.
Starting from single cell suspensions, the Illumina Single Cell 3′ RNA Prep, T100 kit uses particle-templated instant partitions (PIPs) to combine CRISPR-modified cells with barcoded beads to produce sequencing-ready libraries in less than 2 days. These libraries enable 3ʹ gene expression profiling, multiplexed cellular protein measurements, and sample multiplexing of
Illumina Single Cell 3′ RNA and SEA Kit libraries can be pooled together for sequencing, taking into account the differences in cell number and read depth requirements for each library type. Sequencing depth varies based on your application needs, but recommended starting ranges are listed in the following table.
|
sgRNA Reads per Input Cell |
Sequencing Pooling Ratios (sgRNA:Illumina Single Cell 3′ RNA) |
|---|---|
|
1,000–4,000 |
1:20–1:5 |
These libraries require a minimum of 1% PhiX to be added to the loaded library pools. It is recommended to increase PhiX to a minimum of 2% for NovaSeq X Series sequencing instruments.