Overview
This resource explains how to prepare up to 48 uniquely indexed
lllumina Microbial Amplicon Prep—Influenza A/B (IMAP-Flu) offers:
| • | Preparation of ≤ 48 unique dual-indexed libraries |
| • | Scalable library prep from 1–48 samples |
| • | Generation of sequence-ready libraries in < 9 hours |
| • | Assay of Influenza A and Influenza B subtypes |
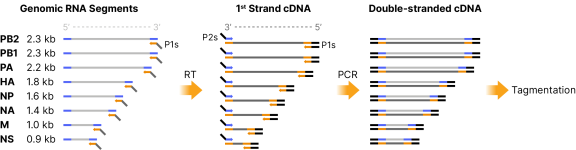
Influenza whole genome RT-PCR approach to generate amplicons ready for tagmentation. The Influenza A/B Primer Pool contains 16 primers designed to hybridize to either the 3’ ends of the genomic RNA segments and 2nd strand cDNA or the 3’ ends of the 1st strand cDNA, enabling both cDNA synthesis of each genomic RNA segment and subsequent PCR amplification to produce whole genome amplicons ready for tagmentation. Blue lines represent the conserved non-coding regions of each genomic RNA segment (or 2nd strand cDNA), the orange lines the complementary portions of the P1 primers, and the black lines represent the additional DNA sequences added by the universal primers to increase primer melting temperatures during PCR.
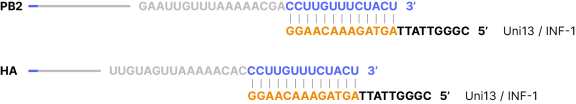
Primer binding site examples for P1 primers to 3’ end of genomic RNAs using the influenza A virus (A/swan/Germany/R65/2006(H5N1)) genomic sequences as target examples. Blue sequences represent complementary sequences of the 3’ ends of the genomic RNAs (or 3’ ends of second strand cDNA) where P1 primers bind.
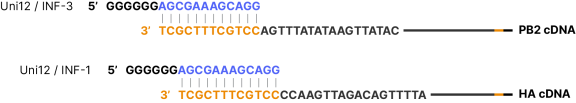
Primer binding site examples for P2 primers binding to the 3’ end of the 1st strand cDNA using influenza A virus (A/swan/Germany/R65/2006(H5N1)) genomic sequences converted to cDNA as target examples.
