Dilute Libraries to the Starting Concentration
This step dilutes libraries to the starting concentration for your sequencing system and is the first step in a serial dilution. After diluting to the starting concentration, libraries are ready to be denatured and diluted to the final loading concentration.
For sequencing, Illumina recommends the read lengths indicated on the Illumina DNA Prep Compatible Products support page.
For sequencing,
Illumina DNA/RNA UD Indexes uses 10 base pair index codes that differ from the Nextera DNA CD Indexes, which use eight base pair index codes. This change in base pair index codes can require adjustments to your sequencing run setup.
| 1. | Calculate the molarity value of the library or pooled libraries using the following formula. |
| • | For libraries qualified on a Bioanalyzer, use the average size obtained for the library. |
| • | For all other qualification methods, use 600 bp as the average library size. |
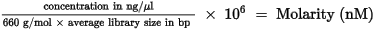
| 2. | Using the molarity value, calculate the volumes of RSB and library needed to dilute libraries to the starting concentration for your system. |
|
Sequencing System |
Starting Concentration (nM) |
Final Loading Concentration (pM) |
|---|---|---|
|
HiSeq 2500 and HiSeq 2000 (high output modes) |
2 |
12 |
|
HiSeq 2500 (rapid run mode) |
2 |
8.5 |
|
HiSeq X, HiSeq 4000, and HiSeq 3000 |
2–3 |
200–300 |
|
iSeq 100 (v1 reagents) |
2 |
200 |
|
iSeq 100 (v2 reagents) |
2 |
100 |
|
MiniSeq |
2 |
1.2–1.3 |
|
MiSeq (v2 and v3 reagents) |
4 |
12 |
|
MiSeq i100* |
0.8 |
80 |
|
NextSeq 550 and NextSeq 500 |
2 |
1.2–1.3 |
|
NextSeq 2000 |
2 |
750 |
|
NovaSeq 6000 |
Refer to the NovaSeq 6000 documentation on the Illumina support site. |
Refer to the NovaSeq 6000 documentation on the Illumina support site. |
|
NovaSeq X |
2 |
150 |
* Denaturation performed onboard. Refer to the system guide.
| 3. | Dilute libraries using RSB: |
| • | Libraries quantified as a multiplexed library pool—Dilute the pool to the starting concentration for your system. |
| • | Libraries quantified individually—Dilute each library to the starting concentration for your system. |
Add 10 µl each diluted library to a tube to create a multiplexed library pool.
| 4. | Follow the denature and dilute instructions for your system to dilute to the final loading concentration. |
| • | The final loading concentrations are a starting point and general guideline. Optimize concentrations for your workflow and quantification method over subsequent sequencing runs or by flow cell titration. |
