Illumina RNA Prep, Tagmentation (L) with Enrichment
Protocol Checklist
| 1. |
|
| 2. | Add 8.5 µl EPH3. |
| 3. | Pipette 10 times. |
| 4. | Centrifuge at 280 × g for 3 seconds. |
| 5. | Place on the thermal cycler and run the DEN_RNA program. |
Total program time is ~5 minutes.
| 6. | Centrifuge at 280 × g for 10 seconds. |
Each well contains 17 µl denatured RNA bound with random hexamers.
| 1. |
|
| • | FSA (9 µl) |
| • | RVT (1 µl) |
Volumes include reagent overage for accurate pipetting.
| 2. | Thoroughly pipette First Strand Synthesis Master Mix. |
| 3. | Add 8 µl First Strand Synthesis Master Mix. |
| 4. | Pipette 10 times. |
| 5. | Centrifuge at 280 × g for 10 seconds. |
| 6. | Place on the thermal cycler and run the FSS program. |
Total program time is ~43 minutes and each well contains a volume of 25 µl.
| 1. | Centrifuge at 280 × g for 10 seconds. |
| 2. | Invert SMM to mix, and then centrifuge briefly. |
| 3. | Add 25 µl SMM. |
| 4. | Pipette 10 times. |
| 5. | Centrifuge at 280 × g for 10 seconds. |
| 6. | Place on the thermal cycler and run the SSS program. |
Total program time is ~1 hour and each well contains a volume of 50 µl.
| 7. | Centrifuge at 280 × g for 10 seconds. |
| 8. | Add 90 µl AMPure XP. |
| 9. |
|
| 10. | Incubate at room temperature for 5 minutes. |
| 11. | Centrifuge at 280 × g for 10 seconds, and then unseal. |
| 12. | Place on the magnetic stand until liquid is clear. |
| 13. | Remove and discard supernatant. |
| 14. | Wash beads as follows. |
| a. |
|
| b. | Wait 30 seconds. |
| c. | Remove and discard supernatant. |
| 15. | Repeat wash a second time. |
| 16. |
|
| 17. | Air-dry for 2 minutes. |
| 18. | Remove from the magnetic stand. |
| 19. | Add 19.5 µl RSB. |
| 20. |
|
| 21. | Incubate at room temperature for 2 minutes. |
| 22. | Centrifuge at 280 × g for 10 seconds, and then unseal. |
| 23. | Place on the magnetic stand until liquid is clear. |
| 24. | Transfer 17.5 µl supernatant. |
Small amounts of bead carryover do not affect performance.
SAFE STOPPING POINT
If you are stopping, seal the plate and store at ‑25°C to ‑15°C for up to 7 days.
| 1. | Centrifuge at 280 × g for 10 seconds. |
| 2. |
|
| • | TB1 (11.5 µl) |
| • | EBLTL (11.5 µl) |
| • | Nuclease-free ultrapure water (14.5 µl) |
Volumes include reagent overage for accurate pipetting.
| 3. | Thoroughly vortex the Tagmentation Master Mix. |
| 4. | Add 32.5 µl Tagmentation Master Mix. |
| 5. | Pipette thoroughly. |
| 6. | Place on the thermal cycler and run the TAG program. |
Total program time is ~5 minutes and each well contains a volume of 50 µl.
| 7. | Centrifuge at 280 × g for 10 seconds. |
| 8. | Incubate at room temperature for 2 minutes. |
| 9. | Add 10 µl ST2. |
| 10. |
|
| 11. | Incubate at room temperature for 5 minutes. |
| 12. | Centrifuge at 280 × g for 10 seconds, and then unseal. |
| 13. | Place on the magnetic stand until liquid is clear. |
| 14. | Remove and discard supernatant. |
| 15. | Wash beads as follows. |
| a. | Remove from the magnetic stand. |
| b. | Add 100 µl TWB to each well. |
| c. |
|
| d. | Centrifuge at 280 × g for 3 seconds. |
| e. | Place on the magnetic stand until liquid is clear. |
| f. | Remove and discard supernatant. |
| 16. | Wash beads a second time. |
| 17. | Wash beads a third time, skipping step f. |
TWB remains in the wells to prevent over-drying.
Each well contains 100 µl beads with tagmented cDNA.
| 18. | Combine the following volumes to prepare PCR Master Mix. |
| • | EPM (23 µl) |
| • | Nuclease-free ultrapure water (23 µl) |
Volumes include reagent overage for accurate pipetting.
| 19. | Thoroughly vortex PCR Master Mix. |
| 20. |
|
| 21. |
|
Foam is normal and does not affect the library.
| 22. | Remove from the magnetic stand. |
| 23. | Add 40 µl PCR Master Mix. |
| 24. |
|
| 25. | Add 10 µl UDP0XXX. |
| 26. |
|
| 27. | Centrifuge at 280 × g for 3 seconds. |
| 28. | Place on the thermal cycler and run the TAG_PCR program. |
Total program time is ~50–60 minutes and each well contains 50 µl beads with DNA attached.
SAFE STOPPING POINT
If you are stopping, seal the plate and store at ‑25°C to ‑15°C for up to 7 days. Alternatively, leave on the thermal cycler for up to 24 hours.
| 1. | Centrifuge at 280 × g for 10 seconds. |
| 2. | Place on the magnetic stand until liquid is clear. |
| 3. | Transfer 45 µl supernatant. |
| 4. | Add 81 µl AMPure XP. |
| 5. |
|
| 6. | Incubate at room temperature for 5 minutes. |
| 7. | Centrifuge at 280 × g for 10 seconds, and then unseal. |
| 8. | Place on the magnetic stand until liquid is clear. |
| 9. | Remove and discard supernatant. |
| 10. | Wash beads as follows. |
| a. |
|
| b. | Wait 30 seconds. |
| c. | Remove and discard all supernatant. |
| 11. | Wash beads a second time. |
| 12. |
|
| 13. | Air-dry on the magnetic stand for 2 minutes. |
| 14. | Remove from the magnetic stand. |
| 15. | Add 17 µl RSB. |
| 16. |
|
| 17. | Incubate at room temperature for 2 minutes. |
| 18. | Centrifuge at 280 × g for 10 seconds, and then unseal. |
| 19. | Place on the magnetic stand until liquid is clear. |
| 20. | Transfer 15 µl supernatant. |
Safe Stopping Point
If you are stopping, seal the plate and store at -25°C to -15°C for up to 30 days.
| 1. | Analyze 1 μl library with the Qubit dsDNA BR Assay Kit. |
| 2. | [Optional] Analyze 1 µl library with the Agilent 2100 Bioanalyzer System and a DNA 1000 Kit. |
| 3. | [RPIP Libraries] |
| • | For one-plex enrichment, dilute one 200 ng library to 7.5 µl. |
| • | For three-plex enrichment, dilute three 200 ng libraries to 2.5 µl each. |
| 4. | [Respiratory Virus Panel Libraries] |
| • | For one-plex enrichment, transfer 7.5 µl undiluted library to one well. |
| • | For three-plex enrichment, dilute three 200 ng libraries to 2.5 µl each. |
| 5. | [All Other Libraries] Dilute libraries in RSB as follows. |
| • | For one-plex enrichment, dilute one 200 ng library to 7.5 µl. |
| • | For three-plex enrichment, dilute three 200 ng libraries to 2.5 µl each. |
| 6. | [Diluted Libraries] In one well, combine the 200 ng libraries: |
|
Number of Libraries |
Total Mass (ng) |
Total Volume (µl) |
|---|---|---|
|
1 |
200 |
7.5 |
|
3 |
600 |
7.5 |
| • | If the total volume is > 7.5 µl, concentrate the pooled sample to 7.5 µl. |
| 1. | Add the following volumes in the order listed. |
| • | 200 ng library or 600 ng pool (7.5 µl) |
| • | NHB2 (12.5 µl) |
| • | Enrichment oligos (2.5 µl) |
| • | EHB2 (2.5 µl) |
| 2. | Pipette 10 times to mix. |
| 3. | Centrifuge at 280 × g for 3 seconds. |
EHB2 can make the reaction appear cloudy, which is normal.
| 4. | Place on the thermal cycler and run the HYB program. |
| 5. |
|
Each well contains a volume of 25 µl.
| 1. |
|
| • | EE1 (28.5 µl) |
| • | HP3 (1.5 µl) |
Volumes include reagent overage for accurate pipetting despite possible foam.
| 2. | Thoroughly pipette Elution Master Mix, and then set aside. |
| 3. | Centrifuge the PCR plate at 280 × g for 10 seconds. |
| 4. | Add 62.5 µl SMB3. |
| 5. |
|
| 6. | Place in the 58°C thermal cycler for 15 minutes. |
The thermal cycler runs continuously through the capture and four washes.
| 7. | Immediately do as follows. |
| a. | Centrifuge at 280 × g for 10 seconds. |
| b. | Place on the magnetic stand until liquid is clear. |
| 8. | Remove and discard supernatant. |
| 9. | Remove from the magnetic stand. |
| 10. | Add 50 µl preheated EEW. |
| 11. |
|
| 12. | Return unused EEW to the microheating system. |
| 13. | Return the plate to the 58°C thermal cycler for 5 minutes. |
| 14. | Immediately do as follows. |
| a. | Centrifuge at 280 × g for 3 seconds. |
| b. | Place on the magnetic stand until liquid is clear. |
| 15. | Remove and discard supernatant. |
| 16. | Remove from the magnetic stand. |
| 17. | Add 50 µl preheated EEW. |
| 18. |
|
| 19. | Return unused EEW to the microheating system. |
| 20. | Return the plate to the 58°C thermal cycler for 5 minutes. |
| 21. | Immediately do as follows. |
| a. | Centrifuge at 280 × g for 3 seconds. |
| b. | Place on the magnetic stand until liquid is clear. |
| 22. | Remove and discard supernatant. |
| 23. | Repeat steps 16–22. |
| 24. | Remove from the magnetic stand. |
| 25. | Add 50 µl preheated EEW. |
| 26. |
|
| 27. | Centrifuge at 280 × g for 3 seconds. |
| 28. | Transfer 50 µl resuspended bead solution. |
The transfer minimizes residual reagents that can inhibit amplification.
| 29. | Seal and centrifuge at 280 × g for 3 seconds. |
| 30. | Return to the 58°C thermal cycler for 5 minutes. |
| 31. | Immediately place on the magnetic stand until liquid is clear. |
| 32. | Remove and discard supernatant. |
| 33. |
|
| 34. | Combine the following volumes to prepare Elution Master Mix. |
| • | EE1 (28.5 µl) |
| • | HP3 (1.5 µl) |
Volumes include reagent overage for accurate pipetting despite possible foam.
| 35. | Thoroughly pipette Elution Master Mix, and then set aside. |
| 36. | Remove from the magnetic stand. |
| 37. | Add 23 µl Elution Master Mix. |
| 38. |
|
| 39. | Incubate at room temperature for 2 minutes. |
| 40. | Centrifuge at 280 × g for 10 seconds, and then unseal. |
| 41. | Place on the magnetic stand until liquid is clear. |
| 42. | Transfer 21 µl supernatant. |
| 43. | Add 4 µl ET2. |
| 44. |
|
Each well contains a volume of 25 µl.
SAFE STOPPING POINT
If you are stopping, seal the plate and store at ‑25°C to ‑15°C for up to 7 days.
| 1. | Centrifuge the sealed plate at 280 × g for 10 seconds. |
| 2. | Add 5 µl PPC. |
| 3. | Add 20 µl EPM. |
| 4. |
|
| 5. | Centrifuge at 280 × g for 10 seconds. |
| 6. | Place on the thermal cycler and run the AMP program. |
Total program time is ~35 minutes and each well contains a volume of 50 µl.
Safe Stopping Point
If you are stopping, store at 2°C to 8°C for up to 2 days. Alternatively, leave on the thermal cycler for up to 24 hours.
| 1. | Centrifuge at 280 × g for 10 seconds. |
| 2. | Add 90 µl AMPure XP. |
| 3. |
|
| 4. | Incubate at room temperature for 5 minutes. |
| 5. | Centrifuge at 280 × g for 10 seconds, and then unseal. |
| 6. | Place on the magnetic stand until liquid is clear. |
| 7. | Remove and discard supernatant. |
| 8. | Wash beads as follows. |
| a. |
|
| b. | Wait 30 seconds. |
| c. | Remove and discard supernatant. |
| 9. | Wash beads a second time. |
| 10. |
|
| 11. | Air-dry on the magnetic stand for 2 minutes. |
| 12. | Remove from the magnetic stand. |
| 13. | Add 32 µl RSB. |
| 14. |
|
| 15. | Incubate at room temperature for 2 minutes. |
| 16. | Centrifuge at 280 × g for 10 seconds, and then unseal. |
| 17. | Place on the magnetic stand until liquid is clear. |
| 18. | Transfer 30 µl supernatant. |
SAFE STOPPING POINT
If you are stopping, seal the plate and store at ‑25°C to ‑15°C for up to 7 days.
| 1. | Check the enriched library: |
| • | Analyze 1 µl enriched library with the Qubit dsDNA HS Assay kit. |
| • | Analyze 1 µl enriched library with the Agilent 2100 Bioanalyzer System and a DNA 1000 Kit. |
Example Yield From Bioanalyzer
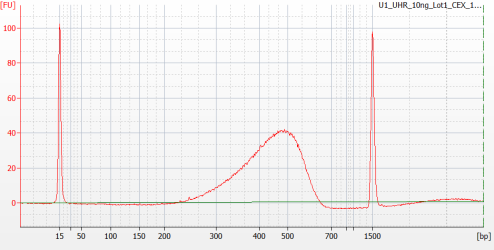
| 1. | Obtain the molarity value: |
| • | Bioanalyzer quantification only—Use the molarity value obtained for the library. |
| • | Bioanalyzer and Qubit quantification—Calculate molarity value using the average size and concentration. |
| 2. | Using the molarity value, calculate the volumes of RSB and library needed to dilute libraries to the starting concentration for your system. |
|
Sequencing System |
Starting Concentration (nM) |
Final Loading Concentration (pM) |
|---|---|---|
|
NextSeq 550 and NextSeq 500 |
20 |
0.8 |
|
NovaSeq 6000 |
0.6 |
120 |
| 3. | Dilute each library to the starting concentration. Combine 10 µl each diluted library in a tube. |
| 4. | Follow denature and dilute instructions to dilute libraries. |
