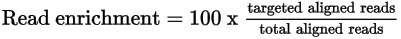Prepare for Pooling
The index adapter plates contain a unique combination of dual i5 and i7 indexes per well. There are a total of 96 unique dual (UD) index combinations per plate. Index adapter plates from sets A and B offer different UD index combinations per set. For index adapter sequences, refer to Illumina Adapter Sequences.
For additional recommendations to plan indexing and pooling strategies for library prep, refer to Index Adapters Pooling.
Illumina Cell-Free DNA Prep with Enrichment reagents are configured and tested at 1-plex and 4-plex enrichment plexity.
| • | Enrichment plexity —The number of pre-enriched libraries pooled together in one enrichment reaction for hybridization with the enrichment probe panel. For example, combining four pre-enriched libraries together creates a 4-plex enrichment pool. |
| • | Enrichment reaction —The number of unique enrichment reaction preparations, regardless of the number of pre-enriched libraries pooled per reaction. For example, a single enrichment reaction can prepare a 1-plex or 4-plex enrichment pool. |
Pre-enriched libraries are pooled during the hybridize probes procedure. 1-plex library and 4-plex library pools are processed in the same manner through the enrichment steps and normalization workflows.
To calculate the total number of enriched libraries, multiply the enrichment plexity per reaction by the number of enrichment reactions. For example, a single enrichment reaction of a 4-plex enrichment pool produces a pool of four enriched libraries.
Enriched libraries are pooled for sequencing after the library normalization step. Libraries should be pooled with nonoverlapping indexes. Each sequenced library should achieve at least 30,000x on-target coverage. Refer to the denature and dilute instructions for your system to calculate library plexity for sequencing and to confirm that each sample achieves sufficient on-target coverage.
Read Enrichment Recommendations
It is recommended that libraries achieve ≥ 60% read enrichment. Libraries with % read enrichment below 60% may require panel optimization or a reduction in sequencing plexity to ensure adequate coverage.
| • | Targeted aligned reads—The number of reads that align to the target. |
| • | Total mapped reads—The total number of pass-filter reads in the data set that aligned to the reference genome. |
Calculate read enrichment using the following formula.