Overview
This guide explains how to prepare up to 384 dual-indexed paired-end libraries from DNA using the Nextera XT DNA Library Prep workflow.
The Nextera XT workflow:
| • | Uses tagmentation, an enzymatic reaction, to fragment DNA and add partial adapter sequences in only 15 minutes. |
| • | Reduces reagent containers, pipetting, and hands-on time using master mixed reagents. |
| • | Requires only 1 ng input DNA. |
| • | Supports genomes that are less than 5 Mb. |
|
Nextera XT (FC-131-1024, FC-131-1096) |
Illumina DNA Prep (20018704, 20018705) |
|---|---|
|
Small genomes, amplicons, plasmids |
Human genomes, large or complex genomes |
|
PCR amplicons (> 300 bp)* |
Small genomes, microbial genomes, plasmids, PCR amplicons (> 150 bp) |
|
Plasmids |
Nonhuman mammalian genomes (eg, mouse, rat, bovine) |
|
Microbial genomes (eg, Prokaryotes, Archaea) |
Plant genomes (eg, Arabidopsis, maize, rice) |
|
Concatenated amplicons |
Invertebrate genomes (eg, Drosophila) |
|
Double-stranded cDNA |
– |
|
Single-cell RNA-Seq |
– |
* Using > 300 bp amplicon size ensures even coverage across the length of the DNA fragment. For more information, refer to PCR Amplicons.
The Nextera XT DNA Library Prep uses an engineered transposome to tagment genomic DNA, which is a process that fragments DNA and then tags the DNA with adapter sequences in one step. Limited-cycle PCR uses the adapters to amplify the insert DNA. The PCR step also adds index adapter sequences on both ends of the DNA, which enables dual-indexed sequencing of pooled libraries on Illumina sequencing platforms.
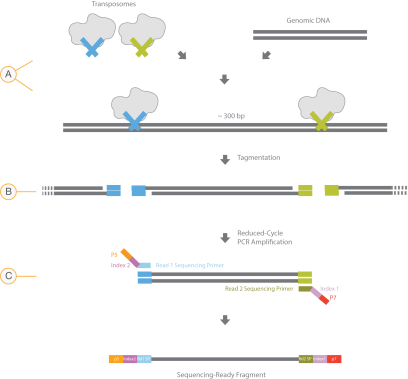
| A. | Nextera XT transposome with adapters combined with template DNA |
| B. | Tagmentation to fragment and add adapters |
| C. | Limited-cycle PCR to add index adapter sequences |
