Introduction
The Illumina Single Cell 3′ RNA Prep, T20 kit uses particle-templated instant partitions (PIPs) to combine single cell or nuclei suspensions with barcoded beads to produce sequencing-ready libraries in less than two days. The assay supports
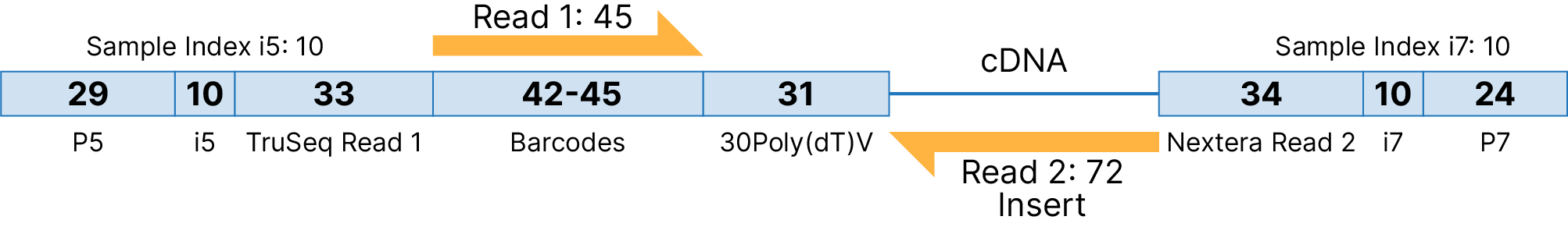
Illumina Single Cell 3′ RNA Prep, T20 kit libraries are composed of standard Illumina paired-end constructs that begin with P5 and end with P7. Read 1 contains the barcode information while Read 2 contains the gene expression information. These libraries are dual-indexed with 10-base i5 and i7 indexes. Read 1 length must be ≥ 45 bases and the recommended Read 2 length ≥ 72 bases.
Illumina Single Cell 3′ RNA Prep libraries can be pooled for sequencing, taking into account the differences in cell number and read depth requirements. Sequencing depth varies based on your application needs but it is recommended to start with a depth of 20,000 reads per input cell. Samples must have distinct index combinations to avoid failures in sample demultiplexing. Refer to Index Adapters Pooling Guide for discussion of appropriate color balance combinations for the selected sequencing platform. After the Illumina Single Cell 3′ RNA Prep libraries are quantified and normalized, dilute and prepare the libraries as recommended in Denature and Dilute Protocol Generator for the selected Illumina sequencing system.
