Nucleotide Diversity
Illumina bases cluster density recommendations on diverse libraries. Diversity describes the proportion of each nucleotide (A, T, C, and G) at each position in a library. Low-diversity libraries compromise software performance and data accuracy.
A balanced or diverse library has equal proportions of A, C, G, and T. Low-diversity libraries, such as PCR amplicon, metagenomic, and ChIP, have an uneven proportion of nucleotides across the flow cell from one cycle to the next. Unbalanced libraries, such as bisulfite-converted libraries, have one base at a much lower percentage than the others.
Cluster Images and Data by Cycle
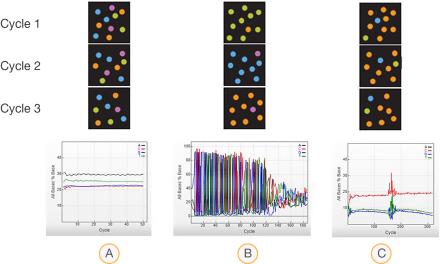
- Diverse libraries—Equal proportions of A, C, T, and G with even, horizontal curves centered on 25%.
- Low-diversity libraries—Uneven proportions of A, C, T, and G with large intensity spikes at each cycle.
- Unbalanced libraries—A low percentage of A and a high percentage of C.
When sequencing low-diversity libraries, use the following strategies to increase nucleotide diversity and provide a balanced signal:
| • | Reduce the loading concentration of the library. Empirically determine the reduction amount. |
| • | Spike in PhiX or other high-diversity library. Empirically determine the spike-in amount, using the following percentages as a general guideline. Start with a higher spike-in percentage and reduce based on run performance. |
|
System |
PhiX Spike-In for Low Diversity (%) |
|---|---|
|
HiSeq 2500 |
≥ 10 |
|
HiSeq 3000 and HiSeq 4000 |
5–20 |
|
HiSeq X |
5–20 |
|
iSeq 100 |
≥ 5 |
|
MiniSeq |
10–50 |
|
MiSeq |
≥ 5 |
|
NextSeq 500 and NextSeq 550 |
10–50 |
|
NovaSeq 6000 |
~5 |
Although providing a balanced signal is not a concern for high-diversity libraries, Illumina recommends a 1–2% PhiX spike-in as a positive control for sequencing.
If a library is new or the nucleotide diversity is unknown, target a conservative loading concentration. A conservative loading concentration is at the lower end of the recommended cluster density range.
A 1–2% PhiX spike-in is also recommended.
