Dual-Indexed Workflow on a Paired-End Flow Cell
Dual-index sequencing on a paired-end flow cell follows one of two workflows, depending on the system and software:
| • | The forward strand workflow is performed on the MiniSeq with Rapid Reagent kits, and the MiSeq. |
| • | The reverse complement workflow is performed on the iSeq 100, MiniSeq with Standard reagent kits, NextSeq Systems, NovaSeq 6000 with v1.5 reagent kits, and NovaSeq X. |
The chemistry applied to the Index 2 Read during a paired-end, dual-indexed run on the MiniSeq with rapid reagent kits, or MiSeq, specific to the paired-end flow cell. Reading the i5 index requires seven additional chemistry-only cycles. This step uses the resynthesis mix, a paired-end reagent, during the Index 2 Read process.
While MiniSeq Rapid reagents allow for dual indexing, Read 2 cannot be performed with the MiniSeq Rapid reagent kit.
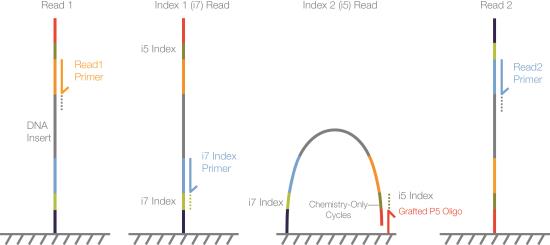
| 1. | Read 1—Read 1 follows the standard Read 1 sequencing protocol using SBS reagents. The Read 1 sequencing primer is annealed to the template strand during the cluster generation step. |
| 2. | Index Read preparation—The Read 1 product is removed and the Index 1 (i7) sequencing primer is annealed to the same template strand. |
| 3. | Index 1 (i7) Read—Following Index Read preparation, the Index 1 (i7) Read performs up to 20 cycles of sequencing. |
The maximum number of cycles in each Index Read depends on the system and run parameters.
| 4. | Index 2 (i5) Read—The Index 1 (i7) Read product is removed and the template anneals to the grafted P5 primer on the surface of the flow cell. The run proceeds through an additional seven chemistry-only cycles (no imaging occurs), followed by up to 20 cycles of sequencing. |
| 5. | Read 2 resynthesis—The Index Read product is removed and the original template strand is used to regenerate the complementary strand. The original template strand is then removed to allow hybridization of the Read 2 sequencing primer. |
| 6. | Read 2—Read 2 follows the standard paired-end sequencing protocol using SBS reagents. |
A dual-indexed run on the iSeq 100, MiniSeq with standard reagent kits, NextSeq Systems, NovaSeq 6000 with v1.5 reagent kits, and NovaSeq X System performs the Index 2 Read after Read 2 resynthesis. This workflow requires a reverse complement of the Index 2 (i5) primer sequence compared to the primer sequence used on other Illumina platforms.
The Index 2 sequencing primer is part of the dual-indexing primer mix for the iSeq 100, MiniSeq with standard reagent kits, NextSeq Systems, NovaSeq 6000 with v1.5 reagent kits, and NovaSeq X systems.
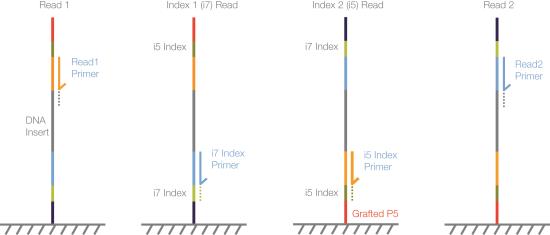
| 1. | Read 1—Read 1 follows the standard Read 1 sequencing protocol using SBS reagents. The Read 1 sequencing primer is annealed to the template strand during the cluster generation step. |
| 2. | Index Read preparation—The Read 1 product is removed and the Index 1 (i7) sequencing primer is annealed to the same template strand. |
| 3. | Index 1 (i7) Read—Following Index Read preparation, the Index 1 (i7) Read performs eight or 10 cycles of sequencing. |
| 4. | Read 2 resynthesis—The Index 1 Read product is removed and the original template strand is used to regenerate the complementary strand. Then the original template strand is removed to allow hybridization of the Index 2 (i5) sequencing primer. |
| 5. | Index 2 (i5) Read—Following Read 2 resynthesis, the Index 2 (i5) Read performs eight or 10 cycles of sequencing. |
This workflow does not require seven additional chemistry-only cycles.
| 6. | Read 2 preparation—The Index 2 Read product is removed and the Read 2 sequencing primer is annealed to the same template strand. |
| 7. | Read 2—Read 2 follows the standard paired-end sequencing protocol using SBS reagents. |
