Illumina MiSeq Integration v6.0 User and Configuration Guide
The Illumina MiSeq Integration Package v6.0 supports the integration of BaseSpace Clarity LIMS to Illumina MiSeq sequencers.
The integration allows for automated tracking of an Illumina sequencing run in the LIMS. This includes tracking instrument run status, generating run report, and capturing and parsing run statistics. In addition, this integration provides automated generation of a sample sheet file for use with the MiSeq Control Software (MCS).
| • | This guide describes the integration between the LIMS and the Illumina MiSeq system. It includes information about protocols and automations, configuration options, installed components, and rules and constraints. |
| • | For instructions on validating and troubleshooting the Illumina MiSeq Integration Package, see the Illumina MiSeq Integration Validation and Troubleshooting Guide. |
| • | For overview information about how the Illumina – LIMS interface works, and which events are monitored, see How the Illumina Instrument and LIMS Software Interact. |
The Illumina MiSeq Integration Package v6.0 includes:
| • | Preconfigured MiSeq Sequencing v1.0 workflow that maps to lab protocol steps and instrument runs. |
| • | Automated generation of a sample sheet for use with the MiSeq Control Software (MCS).Automated tracking of the MiSeq sequencing run and parsing of run statistics into the LIMS, including:Run status and metrics of sequencing run |
| – | Sequencing run parameters |
| – | Real Time Analysis v1 (RTA) run directory location and other run specific information |
Information on these features is provided in this guide.
Note that this integration also includes a preconfigured Library Prep Validation v1.0 workflow, used for validation purposes only. The workflow contains a single-step protocol that models the library prep required to produce normalized libraries that are ready for the MiSeq Sequencing v1.0 workflow. For details, see the Illumina MiSeq Integration Validation and Troubleshooting Guide.
It is assumed that samples enter the MiSeq Sequencing v1.0 workflow as normalized libraries, and have had reagent labels attached. That is, before they are assigned to the workflow:
| • | Samples have been accessioned into the LIMS. |
| • | Samples have been run through QC and library prep. |
| • | Samples have been normalized, and the value is captured in a field called Normalized Molarity (nM). |
For details on sample accessioning, see the following articles in the LIMS Documentation (Samples section):
| • | Sample Accessioning |
| • | Using Sample Lists to Upload and Modify Samples |
Samples can be assigned to the MiSeq Sequencing v1.0 workflow automatically (using a routing script), or manually from the Projects and Samples dashboard (see the Assigning Samples to Workflows article).
Workflows, protocols, and steps
The Illumina MiSeq Integration includes the MiSeq Sequencing v1.0 workflow, which contains a single protocol of the same name.
The MiSeq Sequencing v1.0 protocol includes the following steps:
| • | Library Pooling (MiSeq v1.0) |
| • | Denature and Dilute (MiSeq v1.0) |
| • | MiSeq Run (MiSeq v1.0) |
The following image shows the protocol and steps as displayed to the lab scientist working in BaseSpace Clarity LIMS Lab View.
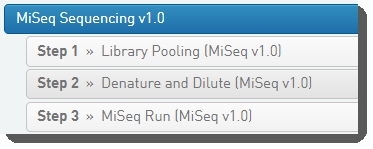
Table 1 lists the step automations configured on the MiSeq Sequencing v1.0 protocol steps. Automation details are provided in the Configuration details sections following the table.
Table 1: MiSeq Sequencing v1.0 protocol step automations
|
Step |
Automation |
||||||
|
1. Library Pooling (MiSeq v1.0) |
|
||||||
|
2. Denature and Dilute (MiSeq v1.0) |
|
||||||
|
3. MiSeq Run (MiSeq v1.0) |
|
The sections below describe the configuration installed with the Illumina MiSeq Integration.
|
Note: The Illumina MiSeq Integration Package v6.0 supports BaseSpace Clarity LIMS v4.2.x and v5.x. As of LIMS v5, the term user-defined field (UDF) has been replaced with custom field in the user interface (the API resource is still called udf). There are two types of custom fields:
|
Step 1: Library Pooling (MiSeq v1.0)
Note that in LIMS v5, the Library Pooling (MiSeq v1.0) step is derived from the Library Pooling v1.0 master step.
In this step, user manually places libraries into pools.
One automation is configured on this step: Set Next Step - Advance
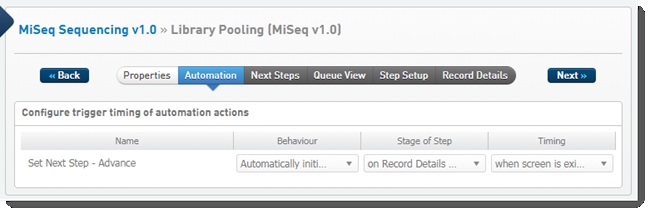 Automation configuration on Library Pooling (MiSeq v1.0) step - LIMS v4.2
Automation configuration on Library Pooling (MiSeq v1.0) step - LIMS v4.2
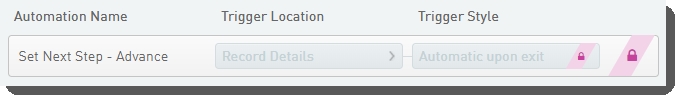 Automation configuration on Library Pooling (MiSeq v1.0) step - LIMS v5 and later
Automation configuration on Library Pooling (MiSeq v1.0) step - LIMS v5 and later
Set Next Step - Advance automation
Automatically triggered on exit of the Record Details screen, this automation advances samples to the next step in the protocol.
Default command line:
bash -l -c "/opt/gls/clarity/bin/java -jar /opt/gls/clarity/extensions/ngs-common/v5/EPP/ngs-extensions.jar -i {stepURI:v2} -u {username} -p {password} \ script:evaluateDynamicExpression \ -t false \ -h false \ -exp 'nextStep = ::ADVANCE::' \ -log {compoundOutputFileLuid0}"
There is one field configured on the Library Pooling (MiSeq v1.0) step. This field displays on the Record Details screen at run time.
The field is shown below in the BaseSpace Clarity LIMS v5 Configuration area (Custom Fields > Master Step Fields tab). See Table 2 for LIMS v4.2 and LIMS v5 field configuration details.
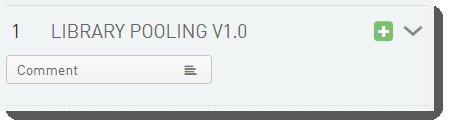
Table 2: Library Pooling (MiSeq v1.0) step UDF / custom field configuration
|
Field Name |
Field Type |
Field Constraints / Options |
||||||
|
Comment |
Multi-line Text |
|
Table 3 lists the global UDFs / fields that are configured to display on the Queue and Ice Bucket screens of the Library Pooling (MiSeq v1.0) step. Note that the majority of these fields display in expanded view only.
Table 3: Global UDF / custom field configuration for Library Pooling (MiSeq v1.0)
|
Field Name |
Field Name |
Field Constraints / Options |
Preset Values / Additional Options and Dropdown Items |
||||||||||||||||||||||||||||||||||||||||||
|
Configured on Sample in LIMS v4.2 and Submitted Samplein LIMS v5: |
|||||||||||||||||||||||||||||||||||||||||||||
|
Application |
Single-line text (v4.2) Text Dropdown (v5) |
|
|
||||||||||||||||||||||||||||||||||||||||||
|
Pooling |
Single-line text (v4.2) Text Dropdown (v5) |
|
|
||||||||||||||||||||||||||||||||||||||||||
|
Read Length |
Single-line text (v4.2) Text (v5) |
|
|||||||||||||||||||||||||||||||||||||||||||
|
Sequencing Coverage |
Single-line text (v4.2) Text (v5) |
|
|||||||||||||||||||||||||||||||||||||||||||
|
Sequencing Method |
Single-line text (v4.2) Text Dropdown (v5) |
|
|
||||||||||||||||||||||||||||||||||||||||||
|
Configured on Analyte in LIMS v4.2 and Derived Samplein LIMS v5: |
|||||||||||||||||||||||||||||||||||||||||||||
|
Normalized Molarity (nM) |
Numeric |
|
|
||||||||||||||||||||||||||||||||||||||||||
| 1. | On the Queue screen: Add the samples to the Ice Bucket. |
| • | In the Add Control Samples panel, add the PhiX v3 control sample to the Ice Bucket. 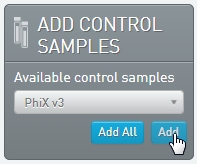 |
| • | Click View Ice Bucket. |
| 2. | On the Ice Bucket screen, click Begin Work. |
| 3. | On the Pool Samples screen: Create a pool of samples by dragging samples into the Pool Creator. |
| • | Name the pool, or accept the default name - Pool #1. 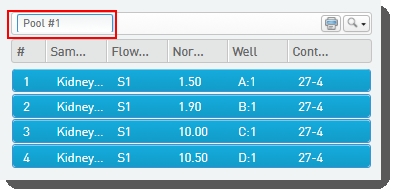 |
| • | Click Place Samples. |
| 4. | On the Placement screen: Select the pool in the Samples to be Placed area on the left and drag it over to the container on the right. |
| • | Click Record Details. |
| 5. | On the Record Details screen, click Next Steps. |
| 6. | On the Assign Next Steps screen, next step is already set to Denature and Dilute (MiSeq v1.0). Click Finish Step. |
Step 2: Denature and Dilute (MiSeq v1.0)
In this step, pooled libraries are denatured and diluted and placed into the reagent cartridge that will be loaded into the MiSeq instrument.
The following automations are configured on this step:
| • | Generate MiSeq SampleSheet |
| • | Set Next Step - Advance |
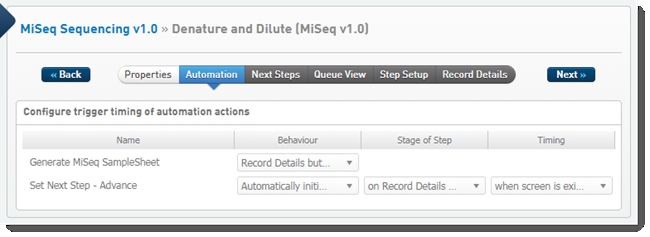 Automation configuration on Denature and Dilute (MiSeq v1.0) step - LIMS v4.2
Automation configuration on Denature and Dilute (MiSeq v1.0) step - LIMS v4.2
 Automation configuration on Denature and Dilute (MiSeq v1.0) step - LIMS v5 and later
Automation configuration on Denature and Dilute (MiSeq v1.0) step - LIMS v5 and later
Generate MiSeq SampleSheet automation
Triggered when the user clicks a button on the Record Details screen, this automation generates generates the sample sheet and attaches it to the step. For details, see Sample sheet generation section, below.
Default command line:
bash -l -c "/opt/gls/clarity/bin/java -jar /opt/gls/clarity/extensions/ngs-common/v5/EPP/DriverFileGenerator.jar -u {username} -p {password} \ script:driver_file_generator \ -i {processURI:v2} \ -t /opt/gls/clarity/extensions/conf/driverfiletemplates/MiSeqSamplesheet.csv \ -o {compoundOutputFileLuid1}.csv \ -q true \ -destLIMSID {compoundOutputFileLuid1} \ -l {compoundOutputFileLuid2}"
Set Next Step - Advance automation
Automatically triggered on exit of the Record Details screen, this automation advances samples to the next step in the protocol.
In LIMS v4.2, the automation also invokes the setUDF script, which sets the value of the Progress UDF. The LIMS updates this UDF value as samples progress through the workflow steps. The Progress UDF does not display in the LIMS user interface, but it does display in the LabLink Collaborations Interface so that collaborators can see the status of their samples.
Default command line (LIMS v4.2):
bash -l -c "/opt/gls/clarity/bin/java -jar /opt/gls/clarity/extensions/ngs-common/v5/EPP/ngs-extensions.jar -u {username} -p {password} \ script:setUDF \ -i {processURI:v2} \ -f 'Progress' \ -t '//input/@uri->//sample/@uri' \ -v 'Library ready for sequencing' \ && /opt/gls/clarity/bin/java -jar /opt/gls/clarity/extensions/ngs-common/v5/EPP/ngs-extensions.jar -u {username} -p {password} \ script:evaluateDynamicExpression \ -i {stepURI:v2} \ -t false \ -h false \ -exp 'nextStep = ::ADVANCE::' \ -log {compoundOutputFileLuid3}"
Default command line (LIMS v5):
bash -l -c "/opt/gls/clarity/bin/java -jar /opt/gls/clarity/extensions/ngs-common/v5/EPP/ngs-extensions.jar -i {stepURI:v2} -u {username} -p {password} \ script:evaluateDynamicExpression \ -t false \ -h false \ -exp 'nextStep = ::ADVANCE::' \ -log {compoundOutputFileLuid3}"
There are 19 fields configured on the Denature and Dilute (MiSeq v1.0) step. These are shown below in the BaseSpace Clarity LIMS v5 Configuration area (Custom Fields > Master Step Fields tab).
At run time, these fields display on the Record Details screen, in the Step Data table. The user manually populates the fields (see User interaction ) and the values are then used to generate the sample sheet. For details on the sample generation functionality, see Sample sheet generation section below.
See Table 4 for LIMS v4.2 and LIMS v5 field configuration details.
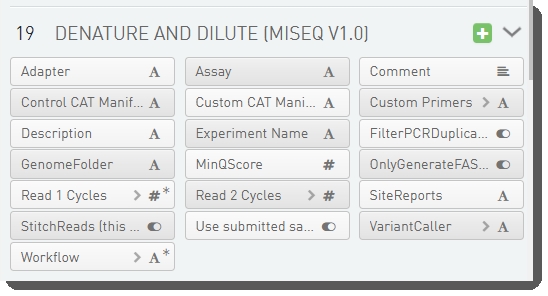
Table 4: Denature and Dilute (MiSeq v1.0) step UDF / custom field configuration
|
Field Name |
Field Type |
Field Constraints / Options |
Preset Values / Additional Options and Dropdown Items |
|||||||||||||||||||||||||||||||||
|
Adapter |
Single-line Text (v4.2) Text (v5) |
|
||||||||||||||||||||||||||||||||||
|
Assay |
Single-line Text (v4.2) Text (v5) |
|
||||||||||||||||||||||||||||||||||
|
Comment |
Multi-line Text |
|
||||||||||||||||||||||||||||||||||
|
Control CAT Manifest |
Single-line Text (v4.2) Text (v5) |
|
||||||||||||||||||||||||||||||||||
|
Custom CAT Manifest |
Single-line Text (v4.2) Text (v5) |
|
||||||||||||||||||||||||||||||||||
|
Custom Primers |
Single-line Text (v4.2) Text Dropdown (v5) |
|
|
|||||||||||||||||||||||||||||||||
|
Description |
Single-line Text (v4.2) Text (v5) |
|
||||||||||||||||||||||||||||||||||
|
Experiment Name |
Single-line Text (v4.2) Text (v5) |
|
||||||||||||||||||||||||||||||||||
|
FilterPCRDuplicates |
Check Box (v4.2) Toggle Switch (v5) |
|
|
|||||||||||||||||||||||||||||||||
|
GenomeFolder |
Single-line Text (v4.2) Text (v5) |
|
||||||||||||||||||||||||||||||||||
|
MinQScore |
Numeric |
|
|
|||||||||||||||||||||||||||||||||
|
OnlyGenerateFASTQ |
Check Box (v4.2) Toggle Switch (v5) |
|
|
|||||||||||||||||||||||||||||||||
|
Read 1 Cycles |
Numeric (v4.2) Numeric Dropdown (v5) |
|
|
|||||||||||||||||||||||||||||||||
|
Read 2 Cycles |
Numeric (v4.2) Numeric Dropdown (v5) |
|
|
|||||||||||||||||||||||||||||||||
|
SiteReportsHidden field, does not display on Record Details screen |
Single-line Text (v4.2) Text (v5) |
|
||||||||||||||||||||||||||||||||||
|
StitchReads(only for GenerateFASTQ, Amplicon-DS and TruSeqAmplicon workflows) |
Check Box (v4.2) Toggle Switch (v5) |
|
|
|||||||||||||||||||||||||||||||||
|
Use submitted sample details for Genome Folder location |
Check Box (v4.2) Toggle Switch (v5) |
|
|
|||||||||||||||||||||||||||||||||
|
VariantCaller |
Single-line Text (v4.2) Text Dropdown (v5) |
|
|
|||||||||||||||||||||||||||||||||
|
Workflow |
Single-line Text (v4.2) Text Dropdown (v5) |
|
|
The following group of defaults / presets are configured for Denature and Dilute (MiSeq v1.0) step UDFs / master step custom fields:
Resequencing
| • | Custom Primers: None |
| • | FilterPCRDuplicates: Yes |
| • | GenomeFolder: Required |
| • | OnlyGenerateFastQ: No |
| • | Read 1 Cycles: 251 |
| • | StitchReads: None Set |
| • | Use submitted sample details for Genome Folder location: No |
| • | Workflow: Resequencing |
Custom Amplicon
| • | Custom CAT Manifest: Required |
| • | Custom Primers: None |
| • | FilterPCRDuplicates: Yes |
| • | GenomeFolder: Required |
| • | OnlyGenerateFastQ: No |
| • | Read 1 Cycles: 251 |
| • | StitchReads: None Set |
| • | Use submitted sample details for Genome Folder location: No |
| • | Workflow: Custom Amplicon |
PCR Amplicon
| • | Custom CAT Manifest: Required |
| • | Custom Primers: None |
| • | FilterPCRDuplicates: Yes |
| • | GenomeFolder: Required |
| • | OnlyGenerateFastQ: No |
| • | Read 1 Cycles: 251 |
| • | StitchReads: None Set |
| • | Use submitted sample details for Genome Folder location: No |
| • | Workflow: PCR Amplicon |
Enrichment
| • | Custom CAT Manifest: Required |
| • | Custom Primers: None |
| • | FilterPCRDuplicates: Yes |
| • | GenomeFolder: Required |
| • | OnlyGenerateFastQ: No |
| • | Read 1 Cycles: 251 |
| • | StitchReads: None Set |
| • | Use submitted sample details for Genome Folder location: No |
| • | Workflow: Enrichment |
Library QC
| • | Custom Primers: None |
| • | FilterPCRDuplicates: Yes |
| • | GenomeFolder: Required |
| • | OnlyGenerateFastQ: No |
| • | Read 1 Cycles: 251 |
| • | StitchReads: None Set |
| • | Use submitted sample details for Genome Folder location: No |
| • | Workflow: LibraryQC |
Metagenomics
| • | Custom Primers: None |
| • | FilterPCRDuplicates: Yes |
| • | OnlyGenerateFastQ: No |
| • | Read 1 Cycles: 251 |
| • | StitchReads: None Set |
| • | Use submitted sample details for Genome Folder location: No |
| • | Workflow: Metagenomics |
GenerateFastQ
| • | Custom Primers: None |
| • | FilterPCRDuplicates: Yes |
| • | OnlyGenerateFastQ: Yes |
| • | Read 1 Cycles: 251 |
| • | StitchReads: None Set |
| • | Use submitted sample details for Genome Folder location: No |
| • | Workflow: GenerateFastQ |
Assembly
| • | Custom Primers: None |
| • | FilterPCRDuplicates: Yes |
| • | OnlyGenerateFastQ: No |
| • | Read 1 Cycles: 251 |
| • | StitchReads: None Set |
| • | Use submitted sample details for Genome Folder location: No |
| • | Workflow: Assembly |
Table 5 shows the global UDFs / fields that are configured to display on the Queue, Ice Bucket and Record Details screens of the Denature and Dilute (MiSeq v1.0) step.
Table 5: Global UDF / custom field configuration for Denature and Dilute (MiSeq v1.0) step
|
Field Name |
Field Type |
Field Constraints / Options |
Preset Values / Additional Options and Dropdown Items |
|||||||||||||||||||||
|
Configured on Sample in LIMS v4.2 and Submitted Samplein LIMS v5: |
||||||||||||||||||||||||
|
Progress(LIMS v4.2) Displays in LabLink Collaborations Interface only |
Single-line Text (v4.2) Text (v5) |
|
||||||||||||||||||||||
|
Read Length Displays on Queue and Ice Bucket screens |
Single-line Text (v4.2) Text (v5) |
|
||||||||||||||||||||||
|
Sequencing Method Displays on Queue and Ice Bucket screens |
Single-line Text (v4.2) Text Dropdown (v5) |
|
|
|||||||||||||||||||||
|
Configured on Analyte in LIMS v4.2 and Derived Samplein LIMS v5: |
||||||||||||||||||||||||
|
Final Loading Concentration Displays on Record Details screen |
Numeric (v4.2) Numeric Dropdown (v5) |
|
|
|||||||||||||||||||||
Placeholders for the following files are configured on the Record Details screen of the Denature and Dilute (MiSeq v1.0) step:
| • | Lab Tracking Form (manually generated) |
| • | MiSeq Samplesheet (auto-generated) |
| • | MiSeq Samplesheet Generation Log (auto-generated) |
| • | Log File (auto-generated) |
Table 6: Sample sheet files and placeholders generated by Denature and Dilute (MiSeq v1.0) step
|
Item |
Description |
|
Lab Tracking Formplaceholder |
This placeholder in the LIMS allows the user to manually attach a lab-specific tracking form to the step. |
|
MiSeq Sample Sheet |
This CSV file is automatically generated by the LIMS for use with the MiSeq instrument. May be opened as a text file, or as an MS Excel spreadsheet. |
|
Sample Sheet Generation Log |
Automatically generated by the LIMS, this log file captures any errors that the LIMS may encounter when generating the sample sheet. |
|
Log File |
Automatically generated by the LIMS, this log file captures the status of the EvaluateDynamicExpression script that is invoked by the Set Next Step - Advanceautomation. |
| 1. | On the Queue screen, add the pool to the Ice Bucket and click View Ice Bucket. |
| 2. | On the Ice Bucket screen, click Begin Work. |
| 3. | On the Placement screen: Scan the barcode of the MiSeq reagent cartridge into the MiSeq Reagent Cartridge field. |
| • | Place the pool of samples into the reagent cartridge. |
| • | Click Record Details. |
| 4. | On the Record Details screen: Under Reagent Lot Tracking, select the reagent lot used in the step. (You may need to add/activate the lot on the Reagents and Controlsscreen.) 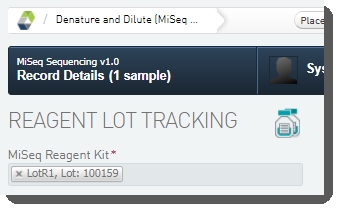 |
| • | Populate the fields in the Step Details table, as appropriate. Note that Workflow and Read 1 Cycles are required fields. |
| • | In the Sample Details table, enter the Final Loading Concentration. You may select from the two preset options - 225 (for PCR-free workflows) or400(for Nano workflows), or enter a different value.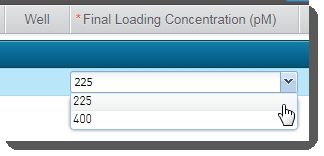 |
| • | Click Generate MiSeq SampleSheet. |
| • | The LIMS generates the samplesheet and attaches it and a log file to placeholders in the Files area of the Record Detailsscreen. Download the files and validate their format and content. |
| • | Click Next Steps. |
| 5. | On the Assign Next Steps screen, samples are already assigned to the MiSeq Run (MiSeq v1.0)step. 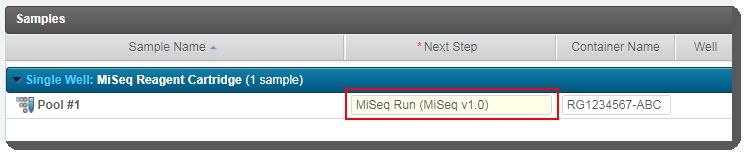 Click Finish Step. Click Finish Step. |
See also Sample sheet generation section.
Step 3: MiSeq Run (MiSeq v1.0)
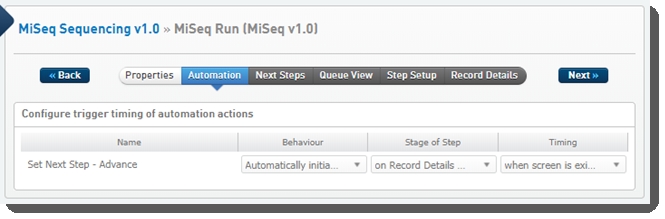
One automation is configured on this step: Set Next Step - Advance
 Automation configuration on MiSeq Run (MiSeq v1.0) step - LIMS v4.2
Automation configuration on MiSeq Run (MiSeq v1.0) step - LIMS v4.2
 Automation configuration on MiSeq Run (MiSeq v1.0) step - LIMS v5 and later
Automation configuration on MiSeq Run (MiSeq v1.0) step - LIMS v5 and later
Set Next Step - Advance automation
Automatically triggered on exit of the Record Details screen, this automation advances samples to the next step in the protocol.
Default command line:
bash -l -c "/opt/gls/clarity/bin/java -jar /opt/gls/clarity/extensions/ngs-common/v5/EPP/ngs-extensions.jar -i {stepURI:v2} -u {username} -p {password} \ script:evaluateDynamicExpression \ -t false \ -h false \ -exp 'nextStep = ::ADVANCE::' \ -log {compoundOutputFileLuid5}"
There are 18 fields configured on the MiSeq Run (MiSeq v1.0) step. These fields display on the Record Details screen at run time.
Some of the field values are completed by the user running the step, the remaining fields are automatically populated at the end of the run.
The fields are shown below in the BaseSpace Clarity LIMS v5 Configuration area (Custom Fields > Master Step Fields tab). See Table 7 for LIMS v4.2 and LIMS v5 field configuration details.
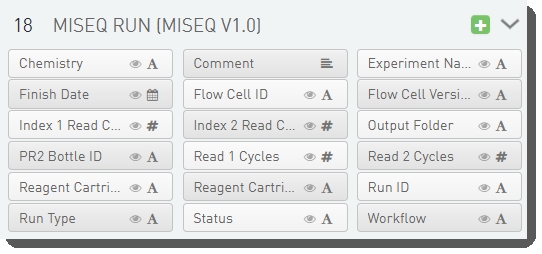
Table 7: MiSeq Run (MiSeq v1.0) step UDF / custom field configuration
|
Field Name |
Field Type |
Field Constraints / Options |
Preset Values / Additional Options and Dropdown Items |
||||||
|
Chemistry |
Single-line Text (v4.2) Text (v5) |
|
|||||||
|
Comment |
Multi-line Text |
|
|||||||
|
Experiment Name |
Single-line Text (v4.2) Text (v5) |
|
|||||||
|
Finish Date |
Date |
|
|||||||
|
Flow Cell ID |
Single-line Text (v4.2) Text (v5) |
|
|||||||
|
Flow Cell Version |
Single-line Text (v4.2) Text (v5) |
|
|||||||
|
Index 1 Read Cycles |
Numeric |
|
Decimal Places Displayed: 0 |
||||||
|
Index 2 Read Cycles |
Numeric |
|
Decimal Places Displayed: 0 |
||||||
|
Output Folder |
Single-line Text (v4.2) Text (v5) |
|
|||||||
|
PR2 Bottle ID |
Single-line Text (v4.2) Text (v5) |
|
|||||||
|
Read 1 Cycles |
Numeric |
|
Decimal Places Displayed: 0 |
||||||
|
Read 2 Cycles |
Numeric |
|
Decimal Places Displayed: 0 |
||||||
|
Reagent Cartridge ID |
Single-line Text (v4.2) Text (v5) |
|
|||||||
|
Reagent Cartridge Part # |
Single-line Text (v4.2) Text (v5) |
|
|||||||
|
Run ID |
Single-line Text (v4.2) Text (v5) |
|
|||||||
|
Run Type |
Single-line Text (v4.2) Text (v5) |
|
|||||||
|
Status |
Single-line Text (v4.2) Text (v5) |
|
|||||||
|
Workflow |
Single-line Text (v4.2) Text (v5) |
|
There are a number of sample and measurement global UDFs / fields that are configured to display on the Record Details screen of the MiSeq Run (MiSeq v1.0) step. These fields are auto-populated at the end of the sequencing run.
For more information, see the User interaction and Sequencing results parsing sections below.
Placeholders for the following files are configured on the Record Details screen of the MiSeq Run (MiSeq v1.0) step:
| • | Illumina Run Report (auto-generated) |
| • | Link to Run Folder (auto-generated) |
| • | Run Parameters (auto-generated) |
| • | Run Info (auto-generated) |
| • | Lab Tracking Form (manually generated) |
| • | Log File (auto-generated) |
For details, see Generated and captured files section below.
| 1. | On the Queue screen, add the pool to the Ice Bucket and click View Ice Bucket. |
| 2. | On the Ice Bucket screen, click Begin Work. |
| 3. | On the Record Details screen, the fields in the table are read-only. Once the run completes, the integration will automatically:Populate the fields. |
| • | Attach files to the Illumina Run Report, Link to Run Folder, Run Parameters, and Run Info placeholders. |
| • | Populate the fields in the Sample Details table. |
| 4. | Click Next Steps. |
| 5. | On the Assign Next Steps screen, the next step is already set to Mark protocol as complete.  Click Finish Step. Click Finish Step. |
Sample sheet generation occurs on the step prior to the sequencing run – Denature and Dilute (MiSeq v1.0), which is the step where samples are placed on the container that will be loaded in the instrument.
The out-of-the-box configuration provides one automation, Generate MiSeq SampleSheet, which uses the Template File Generator (DriverFileGenerator.jar) and a template file to generate a CSV format file for use with the MiSeq Control Software (MCS).
The sample sheet content is determined by the fields that display on the Record Details screen of the step (in the Step Data table), and the values that the user enters into these fields.
You can customize the template used to create the sample sheet, inserting adding additional columns to meet the needs of your lab.
For details:
| • | See the Illumina Instrument Sample Sheets article, which provides a sample template that you can download and customize for your lab. |
| • | For instructions on customizing the template, see the Creating Template Files article. |
The MiSeq Run (MiSeq v1.0) step records information for the flow cell lane and generates a report summarizing the results. In addition, run parameters, run info, and a link to the run folder are automatically captured.
Table 8 describes the run information files, reports, placeholders, and links that the LIMS automatically generates or captures during a sequencing run.
Table 8: Run information generated or captured by MiSeq Run (MiSeq v1.0) step
|
Item |
Description |
|
Run Info Run Parameters |
These XML files are automatically captured by the LIMS from the instrument's run folder. They include the key run parameters, many of which are parsed out into key step UDFs. |
|
Link to Run Folder |
Automatically generated by the LIMS, this is a link to the network run folder where the data that was captured from the instrument during the run is stored. |
|
Illumina Run Report |
Automatically generated by the LIMS, this report provides key information about the run and the samples on the flow cell. Information includes the flow cell ID, run directory location, and primary analysis metrics for the instrument – summarized per flow cell lane for the entire run, and individual reads in the case of paired-end runs. These metrics are compared against the instrument's per lane averages, calculated using metrics from the last 5 sequencing runs. Any values outside of 1 standard deviation are highlighted. |
|
Lab Tracking Form placeholder |
This placeholder in the LIMS allows the user to manually attach a lab-specific tracking form to the step. |
Listed below is the metadata that the LIMS automatically captures from the Illumina sequencing software as part of a sequencing run. This information is gathered from various run result files and events. (To see how these fields are configured in the LIMS, see Table 7.)
| • | Chemistry |
| • | Experiment Name – entered in software |
| • | Finish Date* – run completion date |
| • | Flow Cell ID |
| • | Flow Cell Version |
| • | Index 1 Read Cycles – intended Index cycles |
| • | Index 2 Read Cycles – intended Index cycles |
| • | Output Folder – run folder root |
| • | PR2 Bottle ID |
| • | Reagent Cartridge ID |
| • | Reagent Cartridge Part # |
| • | Read 1 Cycles |
| • | Read 2 Cycles |
| • | Run ID – the unique run ID |
| • | Run Type |
| • | Status – current status of the sequencing run on the instrument |
| • | Workflow |
|
Finish Date is populated as follows:
|
Table 9 lists the Real Time Analysis (RTA) primary analysis metrics the LIMS automatically captures and records, per read, for samples in each flow cell lane. These metrics are captured upon run completion and are stored as UDFs/custom fields in the Record Details screen's Sample Details table.
Users can see both per read and per lane metrics by expanding the output.
Table 9: RTA primary analysis metrics captured by MiSeq Run (MiSeq v1.0) step
|
Per Read LIMS Field Name(stored on derived sample/analyte input to the step) |
Per Lane LIMS Field Name(stored in measurement placeholders in Record Details screen's Sample Details table) |
|
% Aligned R1 |
% Aligned R1 |
|
% Aligned R2 |
% Aligned R2 |
|
% Bases >=Q30 R1 |
% Bases >=Q30 R1 |
|
% Bases >=Q30 R2 |
% Bases >=Q30 R2 |
|
% Error Rate R1 |
% Error Rate R1 |
|
% Error Rate R2 |
% Error Rate R2 |
|
% Phasing R1 |
|
|
% Phasing R2 |
|
|
% Prephasing R1 |
|
|
% Prephasing R2 |
|
|
%PF R1 |
|
|
%PF R2 |
|
|
Cluster Density (K/mm^2) R1 |
|
|
Cluster Density (K/mm^2) R2 |
|
|
Intensity Cycle 1 R1 |
Intensity Cycle 1 R1 |
|
Intensity Cycle 1 R2 |
Intensity Cycle 1 R2 |
|
Reads PF (M) R1 |
|
|
Reads PF (M) R2 |
|
|
Yield PF (Gb) R1 |
Yield PF (Gb) R1 |
|
Yield PF (Gb) R2 |
Yield PF (Gb) R2 |
| 1. | The sequencing service runs on the LIMS server. The service detects event files that the instrument software (RTA) produces as the run progresses; this in turn lets the service know where to find the run data. |
| 2. | As the run data is written out and the End Run event is detected: The data is matched to the step based on the reagent cartridge ID, which was entered/scanned in by the user on the Denature and Dilute (MiSeq v1.0) step. |
| • | Read-only field values on the Record Details screen are populated accordingly. |
| 3. | When it has finished processing the end run event and updating the fields in the LIMS, the sequencing service generates the report and attaches it to the step. |
| 1. | The user manually starts the step and leaves it in progress until the sequencing service has finished processing the run and the run report is generated and attached (see below). |
| 2. | On the Record Details screen, the read-only field values are automatically populated as the sequencer runs. |
| 3. | Once the run has completed:The read-only fields shown in the screenshot above are populated. |
| • | The Illumina Run Report is generated and attached. |
| • | The Link to Run Folder is generated and attached. |
| • | The Run Parameters and Run Info files are attached. |
| 4. | (Optional) In theFiles section, the user uploads the Lab Tracking Form. 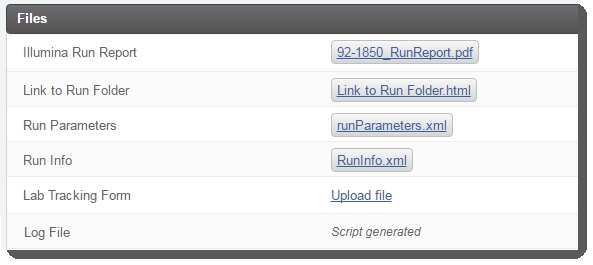 Record Details screen (Files section) of MiSeq Run (MiSeq v1.0) step Record Details screen (Files section) of MiSeq Run (MiSeq v1.0) step |
| 5. | The user completes the step. (Note that the Log File is attached once the next step for samples has been assigned by the Next Step - Advanceautomation.) |
Note that this integration requires installation of the Preconfigured Workflow Package (PCWP). For details, see Release Notes.
The Illumina MiSeq Integration Package v6.0 RPM installs the scripts and files listed in Table 10. Additional component details are provided in the sections following the table.
Table 10: Illumina MiSeq Integration Package v6.0 - scripts and files installed
|
Files Installed |
Location and Description |
|
configure_extensions_miseq_sequencingservice.sh |
/opt/gls/clarity/config/ Script that installs the service properties in the database |
|
log4j.xml |
/opt/gls/clarity/extensions/miseq/SequencingService/conf File containing the settings for the sequencing jar’s logging. |
|
miseq-sequencing.jar |
/opt/gls/clarity/extensions/miseq/SequencingService Jar file containing API-based LIMS extensions used for capturing run results and report generation. |
|
InterOp libraries |
/opt/gls/clarity/extensions/miseq/lib Illumina shared library for parsing InterOp data files. |
|
MiSeqSamplesheet.csv (Installed by the PCWP) |
/opt/gls/clarity/extensions/miseq/Templates Template file used for file generation. |
Support for multiple seqservice.netPathPrefixSearch property values
It is possible to configure support for multiple, identical seqservice.netPathPrefixSearch property values. For details, see Configuring Multiple Identical netPathPrefixSearch Value s.
Table 11 lists the database properties installed with Illumina MiSeq Integration v6.0.
Note that several additional properties, each with the ‘99’ suffix appended to their name, are also installed. These are intended for use by the BaseSpace Clarity LIMS support team in automated validation tests and are not listed in the table.
|
Constraints Sequencing runs are matched using the reagent cartridge ID and the sequencing step’s base name – MiSeq Run (MiSeq v1.0). Do not change this name – it is expected by the sequencing service that captures instrument run results. The base name is stored in the sequenceProcessBaseName database property (see Table 11). If this name is changed without the database property being updated, the 'reagent cartridge ID <-> sequencing step base name' matching system will fail. If required, you may modify the step name by editing or adding to the text after the base name portion as this is not used in the matching system. For example, you could change MiSeq Run (MiSeq v1.0) to MiSeq Run (MiSeq v1.0) v2 |
Table 11: Database properties installed with the Illumina MiSeq Integration Package
|
Property |
Description |
Default Value |
|
miseq.v6.seqservice.sequenceProcessBaseName (installed from the PCWP) |
Sequencing process type / master step base display name. Partial matching is used to look up the process type / master step. |
MiSeq Run (MiSeq v1.0) |
|
miseq.v6.seqservice.eventFileDirectory.1 |
A network location monitored for event files. E.g., /mnt/illumina/gls_events/' |
/mnt/gls_events |
|
miseq.v6.seqservice.netPathPrefixSearch.1 |
The network directory prefix contained in the event file - most likely in Windows format. |
\\nas\network\run_data |
|
miseq.v6.seqservice.netPathPrefixReplace.1 |
The mapped network directory mount name on the server used to access the run data directory. Eg. /mnt/network/data' |
/mnt/run_data |
|
miseq.v6.seqservice.eventFileDirectorySuffixes |
A list of eventFileDirectory path entries to monitor for event files. The value is one or more comma separated integers. |
99 Configured on install to point to "1" |
|
miseq.v6.seqservice.netPathPrefixSearchReplaceSuffixes |
A list of netPathPrefix search and replace entries for transforming Windows to Linux network paths. The value is one or more comma-separated integers. |
99 Configured on install to point to "1" |
|
miseq.v6.seqservice.runReportViewsVersion |
The current version of the Run Report views in the database. The value 0 represents the state before the views are created. This property is automatically updated by the run report. |
0 |
|
miseq.v6.seqservice.ignoreUnmatchedContainerIds |
A flag indicating if event files that cannot be matched to reagent cartridges in the LIMS should be archived after a certain period of time (true), or continually reprocessed (false). |
false Note: To prevent the gls_events file directory from becoming cluttered, it is recommended that the value of this property is set to ‘true’. |
|
miseq.v6.seqservice.ignoreUnmatchedContainerIdsWaitDays |
The number of days between when the event is created and the event file is archived. |
14 |
|
miseq.v6.seqservice.synchronizationPeriod |
Invocation period in seconds. |
60 |
Support for multiple seqservice.netPathPrefixSearch property values
It is possible to configure support for multiple, identical seqservice.netPathPrefixSearch property values. For details, see Configuring Multiple Identical netPathPrefixSearch Values.
Reagent categories/label groups installed
Reagent categories/ label groups are installed with the PCWP workflow configuration slices. For details, see the Reagent Categories / Label Groups article.
The following reagent kits are included in the Illumina MiSeq Integration:
| • | MiSeq Reagent Kit |
The following control types are included in the Illumina MiSeq Integration:
| • | PhiX v3 |
The following container types are included in the Illumina MiSeq Integration:
| • | MiSeq Reagent Cartridge |
See also Support for container types.
|
To ensure that your Illumina instrument warranty remains valid, the instrument integration must be performed and maintained by the BaseSpace Clarity LIMS Support team. To perform this integration, the Support team will require remote access to the instrument while it is idle. |
Below is an overview of the steps performed by the LIMS support team when configuring the instrument for use with the Illumina MiSeq Integration.
Configuring the Illumina instrument:
| 1. | Create directory on the local computer to hold the batch files. These batch files write event files to the network attached storage (NAS) share(s). |
| 2. | Create directory on the NAS to hold the event files. |
| 3. | Modify Illumina software configuration files to call the batch files that create the event files. |
| 4. | Update sequencing service default database properties to match the specifics of the installation. |
All one-dimensional container types with both numeric rows and numeric columns are supported.
This integration operates with the following constraints:
| • | The reagent cartridge ID must be unique. There should not be multiple reagent cartridge containers in the system with the same name. |
| • | The reagent cartridge ID must be scanned as the reagent cartridge Container Name on the Denature and Dilute (MiSeq v1.0) step. |
