Configuration modifications when upgrading from NovaSeq 2.1 or 2.2 to NovaSeq 2.2.1 and NovaSeq 2.2.2
This document is provided for customers who have already installed the Illumina NovaSeq 6000 Integration v2.1or v2.2 packages.
The document shows how to make some minor configuration changes to the NovaSeq 6000 v2.0 workflow to the latest updates.
|
Note:
System Administrator permissions are required to make the configuration changes described below.
|
Part 1: Enabling NovaSeq Xp protocol
Part 1 should be done only when upgrading from Illumina NovaSeq 6000 Integration v2.1 to Illumina NovaSeq 6000 Integration v2.2.1 and v2.2.2
Steps required - LIMS v5
To activate the NovaSeq Xp (NovaSeq 6000 v2.0) protocol for use in the NovaSeq 6000 v2.0 workflow, the following steps are required:
|
1.
|
Display NovaSeq Xp (NovaSeq 6000 v2.0) protocol in Lab View. |
|
2.
|
Configure the Loading Workflow Type global field as follows: Add NovaSeq Xp dropdown list item |
|
•
|
Reorder dropdown list items |
|
•
|
Remove default field value (NovaSeq Standard) configured for NovaSeq v2.1 |
1. Display NovaSeq Xp (NovaSeq 6000 v2.0) protocol in Lab View
|
1.
|
On the Configuration tab, click Lab Work. |
|
2.
|
In the Protocols list, select the NovaSeq Xp (NovaSeq 6000 v2.0) protocol. |
|
3.
|
In the Protocol Settings section, set the Show in Lab View? toggle switch to Yes. |
2. Configure Loading Workflow Type global field
|
1.
|
On the Configuration tab, click Custom Fields. |
|
2.
|
On the Global Fields tab, expand the Derived Samplefield list. |
|
3.
|
Select the Loading Workflow Typefield. In the Field Details area on the right, the dropdown items currently configured for the field are listed under Additional Options. |
|
4.
|
To add a new dropdown item, click the + button, and then type NovaSeq Xpin the text box that displays. As you cannot reorder list items by dragging and dropping them, you now need to add a new item to the end of the list - for the [Remove from workflow] option. You can then delete the existing item. |
|
5.
|
Click inside the [Remove from workflow] text box. Click and drag to select all of the text, and then click Ctrl + Cto copy it to the Clipboard. |
|
6.
|
Click the + button to add another new item. Click inside the text box and then press Ctrl + V to paste the contents of the Clipboard.Because the LIMS does not allow duplicate dropdown list items, both items will be highlighted in red. |
|
7.
|
Click the X button to the right of the original dropdown item to delete it. |
|
8.
|
On the NovaSeq Standard list item, set the Set as Default? toggle switch to No. |
Steps required - LIMS v4.2
To activate the NovaSeq Xp (NovaSeq 6000 v2.0) protocol for use in the NovaSeq 6000 v2.0 workflow, the following steps are required:
|
1.
|
Display NovaSeq Xp (NovaSeq 6000 v2.0) protocol in Lab View. |
|
2.
|
Configure the Loading Workflow Type UDF as follows: Add NovaSeq Xp preset value |
|
•
|
Remove default preset value configured for NovaSeq v2.1 |
1. Display NovaSeq Xp (NovaSeq 6000 v2.0) protocol in Lab View
|
1.
|
In the BaseSpace Clarity LIMS interface, click Configure > Lab. |
|
2.
|
On the Protocols tab, hover over the NovaSeq Xp (NovaSeq 6000 v2.0) protocol and click the Editbutton that displays. |
|
3.
|
On the NovaSeq Xp (NovaSeq 6000 v2.0) configuration screen, clear the Hiddencheck box. |
2. Configure Loading Workflow Type UDF
|
1.
|
In the Operations Interface, on the Configuration menu, click Details > Fields. |
|
2.
|
In the Show Fields for drop-down list, select Analyte. |
|
3.
|
Select the Loading Workflow Type field and click Modify. 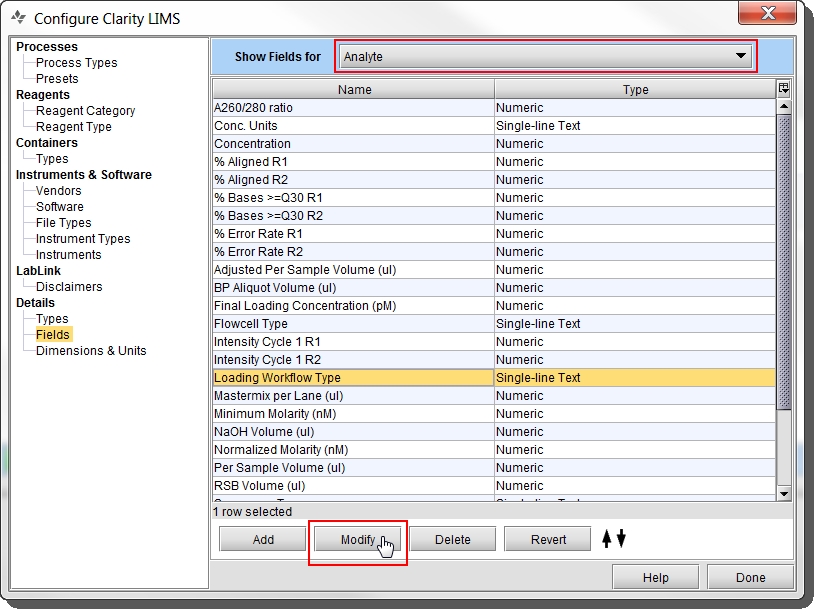 In the Modify Field dialog, the Preset Values tab lists the preset values currently configured for the field. These are the values that display in the Loading Workflow Type drop-down list. In the Modify Field dialog, the Preset Values tab lists the preset values currently configured for the field. These are the values that display in the Loading Workflow Type drop-down list. |
|
4.
|
To add a new value, click the Add button, and then type NovaSeq Xp in the text box that displays. |
|
5.
|
Use the arrow button to move the new value up so that it displays directly below NovaSeq Standard. |
|
6.
|
On the Field Constraints tab, clear the Use first Preset Value as the default valuecheck box. |
Results
The configuration steps described above result in the following changes in the LIMS.
|
•
|
In Lab View The NovaSeq Xp (NovaSeq 6000 v2.0) protocol displays in the Available Workprotocol list. |
|
•
|
When running the Define Run Format (NovaSeq 6000 v2.0) step On the Record Details screen, in the Sample Details table, NovaSeq Xp displays in the Loading Workflow Typedrop-down list. Samples with Loading Workflow Type set to NovaSeq Xp will be routed to the NovaSeq Xp (NovaSeq 6000 v2.0) protocol. |
Part 2: Modifying the Automation for the S1 Flowcell
Part 2 should be done when upgrading from either Illumina NovaSeq 6000 Integration v2.1 or Illumina NovaSeq 6000 Integration v2.2.
Steps required - LIMS v5
|
1.
|
On the Configuration tab, click Automation. |
|
2.
|
On the Step Automation tab, select Calculate Volumes for Make Bulk Pool for NovaSeq Standard. |
|
3.
|
Delete the existing automation and replace with the following: bash -l -c "/opt/gls/clarity/bin/java -jar /opt/gls/clarity/extensions/ngs-common/v5/EPP/ngs-extensions.jar -i {stepURI:v2} -u {username} -p {password} -t true -h false -log {compoundOutputFileLuid1} \script:evaluateDynamicExpression \-exp 'step.::Number of Samples in Pool:: = step.::Number of Samples in Pool:: + 1' \script:evaluateDynamicExpression \-exp 'if ( input.::Flowcell Type:: == ::S1:: ) { step.::Bulk Pool Volume (ul):: = step.::Number of Flowcells to Sequence:: * 100 + 30 } ; if ( input.::Flowcell Type:: == ::S2:: ) { step.::Bulk Pool Volume (ul):: = step.::Number of Flowcells to Sequence:: * 150 + 30 } ; if ( input.::Flowcell Type:: == ::S4:: ) { step.::Bulk Pool Volume (ul):: = step.::Number of Flowcells to Sequence:: * 310 + 30 } ; input.::Per Sample Volume (ul):: = ( ( (input.::Final Loading Concentration (pM):: * 5 / 1000) / input.::Normalized Molarity (nM):: ) * step.::Bulk Pool Volume (ul):: ) / step.::Number of Samples in Pool::' \&& /opt/gls/clarity/bin/java -jar /opt/gls/clarity/extensions/novaseq/novaseq-extensions.jar script:calculate_adjusted_per_sample_volume -i {stepURI:v2} -u {username} -p {password} -l {compoundOutputFileLuid1} \&& /opt/gls/clarity/bin/java -jar /opt/gls/clarity/extensions/ngs-common/v5/EPP/ngs-extensions.jar -i {stepURI:v2} -u {username} -p {password} -t true -h false -log {compoundOutputFileLuid1} \script:evaluateDynamicExpression \-exp 'step.::Total Sample Volume (ul):: = step.::Total Sample Volume (ul):: + input.::Adjusted Per Sample Volume (ul)::' \script:evaluateDynamicExpression \-exp 'if (step.::Total Sample Volume (ul):: >= step.::Bulk Pool Volume (ul)::) {output.::RSB Volume (ul):: = 0} else {output.::RSB Volume (ul):: = step.::Bulk Pool Volume (ul):: - step.::Total Sample Volume (ul)::} ; output.::Flowcell Type:: = input.::Flowcell Type:: ; output.::Loading Workflow Type:: = input.::Loading Workflow Type:: ; if ( input.::Flowcell Type:: == ::S1:: ) { output.::Volume of Pool to Denature (ul):: = 100 ; output.::NaOH Volume (ul):: = 25 ; output.::Tris-HCl Volume (ul):: = 25 } ; if ( input.::Flowcell Type:: == ::S2:: ) {output.::Volume of Pool to Denature (ul):: = 150 ; output.::NaOH Volume (ul):: = 37.50 ; output.::Tris-HCl Volume (ul):: = 37.50 } ; ; if ( input.::Flowcell Type:: == ::S4:: ) { output.::Volume of Pool to Denature (ul):: = 310 ; output.::NaOH Volume (ul):: = 77.50 ; output.::Tris-HCl Volume (ul):: = 77.50 }' \&& /opt/gls/clarity/bin/java -jar /opt/gls/clarity/extensions/ngs-common/v5/EPP/DriverFileGenerator.jar -i {processURI:v2} -u {username} -p {password} -l {compoundOutputFileLuid1} \script:driver_file_generator \-t /opt/gls/clarity/extensions/novaseq/templates/NovaSeq_Standard_Bulk_Pool1.csv \-o 1.csv \script:driver_file_generator \-t /opt/gls/clarity/extensions/novaseq/templates/NovaSeq_Standard_Bulk_Pool2.csv \-o 2.csv \script:driver_file_generator \-t /opt/gls/clarity/extensions/novaseq/templates/NovaSeq_Standard_Bulk_Pool3.csv \-o 3.csv \&& cat 1.csv 2.csv 3.csv > {compoundOutputFileLuid0}.csv" |
Steps required - LIMS v4.2
|
1.
|
In the Operations Interface, on the Configuration menu, click Process > Process Types. |
|
2.
|
In the drop-down list, look for the Make Bulk Pool for NovaSeq Standard (NovaSeq 6000 v2.0) and click Modify. |
|
3.
|
Go to the External Programs tab, select the Calculate Volumes automation and click Modify. |
|
4.
|
Delete the existing automation and replace with the following: bash -l -c "/opt/gls/clarity/bin/java -jar /opt/gls/clarity/extensions/ngs-common/v5/EPP/ngs-extensions.jar -i {stepURI:v2} -u {username} -p {password} -t true -h false -log {compoundOutputFileLuid1} \script:evaluateDynamicExpression \-exp 'step.::Number of Samples in Pool:: = step.::Number of Samples in Pool:: + 1' \script:evaluateDynamicExpression \-exp 'if ( input.::Flowcell Type:: == ::S1:: ) { step.::Bulk Pool Volume (ul):: = step.::Number of Flowcells to Sequence:: * 100 + 30 } ; if ( input.::Flowcell Type:: == ::S2:: ) { step.::Bulk Pool Volume (ul):: = step.::Number of Flowcells to Sequence:: * 150 + 30 } ; if ( input.::Flowcell Type:: == ::S4:: ) { step.::Bulk Pool Volume (ul):: = step.::Number of Flowcells to Sequence:: * 310 + 30 } ; input.::Per Sample Volume (ul):: = ( ( (input.::Final Loading Concentration (pM):: * 5 / 1000) / input.::Normalized Molarity (nM):: ) * step.::Bulk Pool Volume (ul):: ) / step.::Number of Samples in Pool::' \&& /opt/gls/clarity/bin/java -jar /opt/gls/clarity/extensions/novaseq/novaseq-extensions.jar script:calculate_adjusted_per_sample_volume -i {stepURI:v2} -u {username} -p {password} -l {compoundOutputFileLuid1} \&& /opt/gls/clarity/bin/java -jar /opt/gls/clarity/extensions/ngs-common/v5/EPP/ngs-extensions.jar -i {stepURI:v2} -u {username} -p {password} -t true -h false -log {compoundOutputFileLuid1} \script:evaluateDynamicExpression \-exp 'step.::Total Sample Volume (ul):: = step.::Total Sample Volume (ul):: + input.::Adjusted Per Sample Volume (ul)::' \script:evaluateDynamicExpression \-exp 'if (step.::Total Sample Volume (ul):: >= step.::Bulk Pool Volume (ul)::) {output.::RSB Volume (ul):: = 0} else {output.::RSB Volume (ul):: = step.::Bulk Pool Volume (ul):: - step.::Total Sample Volume (ul)::} ; output.::Flowcell Type:: = input.::Flowcell Type:: ; output.::Loading Workflow Type:: = input.::Loading Workflow Type:: ; if ( input.::Flowcell Type:: == ::S1:: ) { output.::Volume of Pool to Denature (ul):: = 100 ; output.::NaOH Volume (ul):: = 25 ; output.::Tris-HCl Volume (ul):: = 25 } ; if ( input.::Flowcell Type:: == ::S2:: ) {output.::Volume of Pool to Denature (ul):: = 150 ; output.::NaOH Volume (ul):: = 37.50 ; output.::Tris-HCl Volume (ul):: = 37.50 } ; ; if ( input.::Flowcell Type:: == ::S4:: ) { output.::Volume of Pool to Denature (ul):: = 310 ; output.::NaOH Volume (ul):: = 77.50 ; output.::Tris-HCl Volume (ul):: = 77.50 }' \&& /opt/gls/clarity/bin/java -jar /opt/gls/clarity/extensions/ngs-common/v5/EPP/DriverFileGenerator.jar -i {processURI:v2} -u {username} -p {password} -l {compoundOutputFileLuid1} \script:driver_file_generator \-t /opt/gls/clarity/extensions/novaseq/templates/NovaSeq_Standard_Bulk_Pool1.csv \-o 1.csv \script:driver_file_generator \-t /opt/gls/clarity/extensions/novaseq/templates/NovaSeq_Standard_Bulk_Pool2.csv \-o 2.csv \script:driver_file_generator \-t /opt/gls/clarity/extensions/novaseq/templates/NovaSeq_Standard_Bulk_Pool3.csv \-o 3.csv \&& cat 1.csv 2.csv 3.csv > {compoundOutputFileLuid0}.csv" |
|
5.
|
Click Save, Save, and Done. |
 In the Modify Field dialog, the Preset Values tab lists the preset values currently configured for the field. These are the values that display in the Loading Workflow Type drop-down list.
In the Modify Field dialog, the Preset Values tab lists the preset values currently configured for the field. These are the values that display in the Loading Workflow Type drop-down list.