Copy Number Variants VCF
File name: {Sample_ID}_cnv.vcf
The copy number variants (CNV) file contains calls for DNA libraries of the CNV genes targeted by TruSight Oncology 500 ctDNA assays. The CNV call indicates fold change results for each gene classified as reference, deletion, or amplification.
The value in the QUAL column of the copy number variants VCF is a Phred transformation of the p-value represented by the following equation:
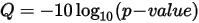
The p-value is derived from the t-test between the fold change (FC) of the gene against the rest of the genome. Higher Q-scores indicate higher confidence in the CNV call.
In the VCF notation, <DUP> indicates the detected FC is greater than a predefined amplification cutoff. <DEL> indicates the FC is less than a predefined deletion cutoff for that gene. This cutoff can vary from gene to gene.
Each copy number variant is reported as the fold change on normalized read depth in a testing sample relative to the normalized read depth in diploid genomes. Given tumor purity, the ploidy of a gene in the sample can be inferred from the reported fold change.
Given tumor purity X%, for a reported fold change Y, the copy number n can be calculated by using the following equation:
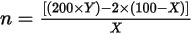
For example, in a testing sample of tumor purity at 30%, MET with a fold change of 2.2x indicates that 10 copies of MET DNA are observed.
