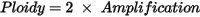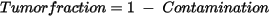HRD Metrics Report
The Illumina DRAGEN TruSight Oncology 500 Analysis Software v2.1.1 analysis software allows for analysis of sequencing data generated from the TruSight Oncology 500 HRD assay. When HRD samples are analyzed new results and metrics are included in the CombinedVariantOutput and MetricsOutput files respectively. The following tables detail how these scores and QC metrics are derived.
|
Metric |
Description |
|---|---|
|
GIS Score |
Proprietary Genomic Instability Score (GIS) indicating level of genomic instability in sample genome. Combination of Loss of Heterozygosity (LOH), Telomeric allelic imbalance and Large-scale State Transitions (LST) scores. The GIS scores provided by TruSight Oncology 500 HRD show good correlation (R2= 0.98) with Myriad Genetics GIS however they are not identical (Refer to TruSight Oncology 500 HRD Product Data Sheet Doc# M-GL-00748 for more details). GIS from alternative HRD assays should be not be considered equivalent to Illumina/Myriad GIS. |
|
Ploidy |
Proprietary estimate of ploidy for the whole genome output. Estimate is very sensitive to noise of b-allele frequencies and has not been validated to assess true genome ploidy. Calculated using “Amplification” in .gis.json file.
|
|
Tumor Fraction |
Proprietary estimate of tumor cell fraction. Estimated may not be accurate for sampleswhich have lower b-allele frequency signals and has not been validated. Calculated from “Contamination“ in .gis.json file. Synthetically generated control samples may also exhibit unexpected behavior.
The “Contamination” used in this calculation and provided in the .gis.json file is NOT the same as the contamination score provided in the MetricsOutput.csv file. These scores are calculated independently. |
|
Metric |
Description |
|---|---|
|
PCT_TARGET_HRD_50X |
Percent of HRD probe SNP panel covered by at least 50X coverage. |
|
ALLELE_DOSAGE_RATIO |
Proprietary Myriad Genetics estimate of b-allele dosage based on b-allele noise/signal ratio. B-Allele noise is correlated with coverage; lower coverage samples will have higher noise. B-allele signal is also correlated with tumor fraction; a higher tumor fraction produces a higher signal for b-allele sites. Samples with lower tumor fraction and higher amount of noise (or lower coverage) will have higher Allele Dosage Ratio. The upper limit of the score is 50, therefore any sample with 50 Allele Dosage Ratio can be assumed to have tumor fraction close to zero and typically has a GIS = 0. |
|
MEDIAN_TARGET_HRD_COVERAGE |
Median target fragment coverage across all target positions in the genome. Coverage is the total number of non-duplicate pair alignments that overlap. |