DRAGEN Methylation Pipeline
The epigenetic methylation of cytosine bases in DNA can have a dramatic effect on gene expression, and bisulfite sequencing is the most common method for detecting epigenetic methylation patterns at single-base resolution. This technique involves chemically treating DNA with sodium bisulfite, which converts unmethylated cytosine bases to uracil, but does not alter methylated cytosines. Subsequent PCR amplification converts any uracils to thymines.
A bisulfite sequencing library can either be nondirectional or directional. For nondirectional, each double-stranded DNA fragment yields four distinct strands for sequencing, post-amplification, as shown in the following figure:
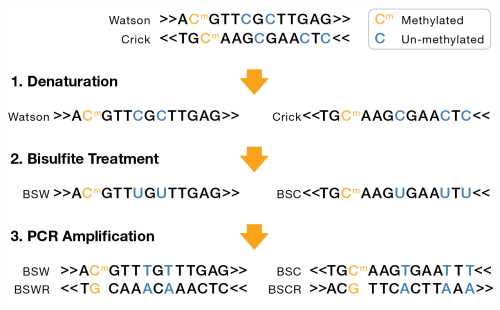
| – | Bisulfite Watson (BSW), reverse complement of BSW (BSWR), |
| – | Bisulfite Crick (BSC), reverse complement of BSC (BSCR) |
For directional libraries, the four strand types are generated, but adapters are attached to the DNA fragments such that only the BSW and BSC strands are sequenced (Lister protocol). Less commonly, the BSWR and BSCR strands are selected for sequencing (eg, PBAT).
BSW and BSC strands:
| • | A, G, T: unchanged |
| • | Methylated C remains C |
| • | Unmethylated C converted to T |
BSWR and BSCR strands:
| • | Bases complementary to original Watson/Crick A, G, T bases remain unchanged. |
| • | G complementary to original Watson/Crick methylated C remains G. |
| • | G complementary to original Watson/Crick unmethylated C becomes A. |
Therefore, several steps are performed to map methylation, as shown in the following flowchart:
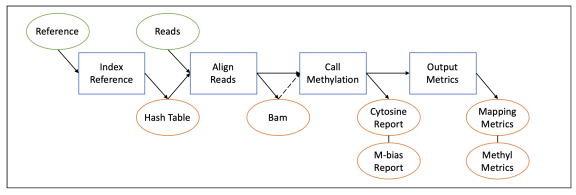
Details on each part of the mapping process can be found in this section.
