DRAGEN Amplicon Pipeline
Amplicon sequencing is a highly targeted approach that enables you to analyze genetic variation in specific genomic regions. The ultradeep sequencing of PCR products (amplicons) allows you to efficiently identify and characterize variants. This method uses oligonucleotide probes designed to target and capture regions of interest, followed by next-generation sequencing (NGS).
The Amplicon Pipeline supports both DNA and RNA data. The Amplicon Pipeline turns off duplicate marking because there are only a few unique start and end postions for fragments from an amplicon target due to the assay.
The DNA Amplicon Pipeline uses the DRAGEN DNA Pipeline by including an additional step after mapping and aligning to soft-clip primers and rewrite alignments. If the target amplicon is found, DRAGEN tags each alignment with the target amplicon and performs soft-clipping the primer sequences. DRAGEN performs tagging by adding an XN:Z:<amplicon name> tag to the output BAM/CRAM record. Soft-clipping makes sure that the primer sequences do not contribute to the variant calls.
![]()
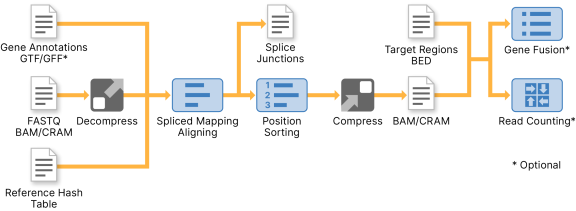
The RNA Amplicon Pipeline uses theDRAGEN RNA Pipeline. Amplicon-specific parameters are set for fusion calling, including a fusion scoring model trained on RNA amplicon data. Small variant calling is not supported in RNA amplicon mode.