Unique Molecular Identifiers
DRAGEN can process data from whole genome and hybrid-capture assays with unique molecular identifiers (UMI). UMIs are molecular tags added to DNA fragments before amplification to determine the original input DNA molecule of the amplified fragments. UMIs help reduce errors and biases introduced by DNA damage such as deamination before library prep, PCR error, or sequencing errors.
To use the UMI Pipeline, the input reads files must be from a paired-end run. Input can be pairs of FASTQ files or aligned/unaligned BAM input.
DRAGEN supports the following UMI types:
| • | Dual, nonrandom UMIs, such as TruSight Oncology (TSO) UMI Reagents or IDT xGen Prism. |
| • | Dual, random UMIs, such as Agilent SureSelect XT HS2 molecular barcodes (MBC) or IDT xGen Duplex Seq Adapters. |
| • | Single-ended, random UMIs, such as Agilent SureSelect XT HS molecular barcodes (MBC) or IDT xGen dual index UMI Adapters. |
DRAGEN uses the UMI sequence to group the read pairs by their original input fragment and generates a consensus read pair for each such group, or family. The consensus reduces error rates to detect rare and low frequency somatic variants in DNA samples with high accuracy. DRAGEN generates a consensus as follows.
| 1. | Aligns reads. |
| 2. | Groups reads into groups with matching UMI and pair alignments. These groups are referred to as families. |
| 3. | Generates a single consensus read pair for each read family. |
These generated reads have higher quality scores than the input reads and reflect the increased confidence gained by combining multiple observations into each base call.
UMI workflow is only compatible with small variant calling and SV in DRAGEN.
Enter UMIs in one of the following formats:
| • | Read name—The UMI sequence is located in the eighth colon-delimited field of the read name (QNAME). For example, NDX550136:7:H2MTNBDXX:1:13302:3141:10799:AAGGATG+TCGGAGA |
| • | BAM tag—The UMI is present as an RX tag in prealigned or aligned BAM file (standard SAM format). |
| • | FASTQ file—The UMI is located in a third FASTQ file using the same read order as the read pairs. |
To create FASTQ, append the UMI to the read name, and then specify the appropriate OverrideCycles setting in the DRAGEN BCL conversion tool (see BCL Data Conversion). DRAGEN supports UMIs with two parts each with a maximum of 8 bp and separated by +, or a single UMI with a maximum of 15 bp.
The UMI workflow must be executed using a set of reads that correspond to a unique set of RGSM/RGLB. DRAGEN supports multiple lanes if all lanes correspond to the same RGSM/RGLB set.
DRAGEN UMI does not support a tumor-normal analysis, because a tumor-normal run corresponds to two different RGSM. In a tumor-normal run, one sample name is used for tumor and one sample name is used for normal. DRAGEN UMI supports one sample in a run.
If using a BAM file or a list of FASTQ files as the input, the input might contain multiple samples. DRAGEN checks if only one sample is included in the run and if the sample uses only a single, unique RGLB library. DRAGEN also accepts a library that was spread across multiple lanes. If there is a single sample and single library, DRAGEN processes all included reads. If there are multiple samples or multiple libraries, DRAGEN aborts analysis with an error.
For dual, nonrandom UMIs, you can provide a predefined UMI correction table or a list of valid UMI sequences as input. To create the UMI correction table, use a tab-delimited file, include a header, and add the following fields.
|
Field |
Value |
|---|---|
|
UMI |
The UMI sequence. For example, ACGTAC. |
|
IsValid |
Specify if the UMI sequence is valid. Enter either: TRUE or FALSE. |
|
NearestCodes |
Colon-separated list of nearest UMI sequences. For example, ACGTAA:ACGTAT. |
|
SecondNearestCodes |
Colon-separated list of second nearest sequences. For example, ACGGAA:ACGGAT. |
If customized correction table is not specified, DRAGEN uses the default table for TruSight Oncology (TSO) UMI Reagents located at src/config/umi_correction_table.txt. Alternatively, you can provide a file for whitelisted nonrandom UMI with valid UMI sequence one per line. DRAGEN then autogenerates a UMI correction table with hamming distance of one.
|
Option |
Description |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
--umi-library-type |
Set the batch option for different UMIs correction. Three batch modes are available that optimize collapsing configurations for different UMI types. Use one of the following modes:
|
|||||||||
|
--umi-min-supporting-reads |
Specify the number of matching UMI inputs reads required to generate a consensus read. Any family with insufficient supporting reads is discarded. For example, the following are the recommended settings for FFPE and ctDNA.
|
|||||||||
|
--umi-enable |
To enable read collapsing, set the --umi-enable option to true. This option is not compatible with –enable-duplicate-marking because the UMI pipeline generates a consensus read from a set of candidate input reads, rather than choosing the best nonduplicate read. If using the --umi-library-type option, --umi-enable is not required. |
|||||||||
|
--umi-emit-multiplicity |
Set the consensus sequence type to output. DRAGEN UMI allows you to collapse duplex sequences from the two strands of the original molecules. Duplex sequence is typically ~20–60% of total library, depending on library kit, input material, and sequencing depth. Enter one of the following consensus sequence types:
|
|||||||||
|
--umi-source |
Specify the input type for the UMI sequence. The following are valid values: qname, bamtag, fastq. If using --umi-source=fastq, provide the UMI sequence from FASTQ file using --umi-fastq. |
|||||||||
|
--umi-correction-table |
Enter the path to a customized correction table. By default, Local Run Manager uses lookup correction with a built-in table for the Illumina TruSight Oncology and Illumina for IDT UMI Index Anchor kits. |
|||||||||
|
--umi-nonrandom-whitelist |
Enter the path for a customized, valid UMI sequence. |
|||||||||
|
--umi-metrics-interval-file |
Enter the path for target region in BED format. |
DRAGEN processes UMIs by grouping reads by UMI and alignment position. If there are sequencing errors in the UMIs, DRAGEN can correct and detect small sequencing errors by using a lookup table or by using sequence similarity and read counts. You specify the type of correction with the --umi-library-type or --umi-correction-scheme option using the values lookup, random, or none.
For sparse sets of nonrandom UMIs, it is possible to create a lookup table that specifies which sequence can be corrected and how to correct it. This correct file scheme works best on UMI sets where sequences have a minimum hamming/edit distance between them. By default, DRAGEN uses lookup correction with a built-in correct table for the Illumina TruSight Oncology and Illumina for IDT UMI Index Anchor kits. Specify the path for your correction file using the --umi-correction-table option. If you are using a different set of nonrandom UMIs, contact Illumina Technical Support for information on generating the corresponding correction file.
In the random UMI correction scheme, DRAGEN must infer which UMIs at a given position are likely to be errors relative to other UMIs observed at the same position. The error modes include small UMI errors, such as one mismatch or UMI jumping or hopping artifact from library prep. DRAGEN accomplishes this as follows.
| • | Groups reads by fragment alignment position. |
| • | Within a small fuzzy window at each position, groups the reads first by the exact UMI sequence, which forms a family. |
| • | Estimate UMI jumping or hopping probability through insert size distribution and number of distinct UMI at certain positions. |
| • | Within a fuzzy window, calculates pair-wise likelihood ratio to assess if two families with different UMI sequences and genomic positions are derived from same original molecule. |
| • | Merges families with likelihood lower than threshold. The default threshold is 1. |
Duplex UMI adapters simultaneously tag both strands of double-stranded DNA fragments. It is then possible to identify reads resulting from amplification of each strand of the original fragment.
DRAGEN considers two collapsed read pairs to be the sequence of two strands of the same original fragment of DNA if they have the same alignment position (within a fuzzy window), complementary orientations, and their UMIs are swapped from Read 1 and Read 2. If there is only single-ended UMI, DRAGEN compares the start-end position of families from two strands and computes pair-wise likelihood to determine if they are likely originated from two distinct families or should be merged as a duplex sequence. By default, DRAGEN outputs both simplex and duplex consensus sequences. To change the consensus sequence output type, use --umi-emit-multiplicity.
Example UMI Commands
The following is an example DRAGEN command for generating a consensus BAM file from input reads with Illumina UMIs:
dragen \
-r <REF> \
-1 <FQ1> \
-2 <FQ2> \
--output-dir <OUTPUT> \
--output-file-prefix <PREFIX> \
--enable-map-align true \
--enable-sort true \
--umi-enable true \
--umi-correction-scheme=lookup \
--umi-min-supporting-reads 2
To run with other random UMI library types, change --umi-library-type to random-simplex or random-duplex.
dragen \
-r <REF> \
-1 <FQ1> \
-2 <FQ3> \
--umi-source=fastq \
--umi-fastq <FQ2> \
--output-dir <OUTPUT> \
--output-file-prefix <
PREFIX> \
--enable-map-align true \
--enable-sort true \
--umi-library-type nonrandom-duplex \
--umi-metrics-interval-file [valid target BED file]
dragen \
-r <REF> \
-1 <FQ1> \
-2 <FQ2> \
--umi-correction-table <valid umi correction table> \
--output-dir <OUTPUT> \
--output-file-prefix <PREFIX> \
--enable-map-align true \
--enable-sort true \
--umi-library-type nonrandom-duplex \
--umi-metrics-interval-file <valid target BED file>
UMI Outputs
If you enable BAM output, DRAGEN generates a <output_prefix>.bam that includes all UMI consensus reads. The QNAMEs for the reads are generated based on the following convention.
consensus_read_refID1_pos1_refID2_pos2_orientation
| • | refID1—The reference ID of Read 1. |
| • | pos1—The genomic position of Read 1. |
| • | refID2—The reference ID of Read 2. |
| • | pos2—The genomic position of Read 2. |
| • | orientation—The orientation of Read 1 and Read 2. Orientation can be one of the following values. Position refers to the outermost aligned position of the read and is adjusted for soft clips. |
| – | 1—Read 1 is forward and Read 2 is reverse. The starting position for Read 1 is less than or equal to the Read 2 end position. |
| – | 2—Read 1 is reverse and Read 2 is forward. The starting position for Read 2 is greater than or equal to the Read 1 end position. |
| – | 3—Read 1 is forward and Read 2 is reverse. The starting position for Read 1 is greater than the Read 2 end position. |
| – | 4—Read 1 is reverse and Read 2 is forward. The starting position for Read 2 is greater than the Read 1 end position. |
| – | 5—Read 1 and Read 2 are forward. |
| – | 6—Read 1 and Read 2 are reverse. |
DRAGEN outputs a <output_prefix>.umi_metrics.csv file that describes the statistics for UMI collapsing. This file summarizes statistics on input reads, how they were grouped into families, how UMIs were corrected, and how families-generated consensus reads. The following metrics can be useful when tuning the pipeline for your application:
| • | Discarded families—Any families having fewer than --umi-min-supporting-reads input or having a different duplex/simplex status than specified by --umi-emit-multiplicity are discarded. These reads are logged as Reads filtered out. The families are logged as Families discarded. |
| • | UMI correction—Families may be combined in various ways. The number of such corrections are reported as follows. |
| – | Families shifted—Families with fragment alignment coordinates up to the distance specified in the umi-fuzzy-window-size parameter. The default umi-fuzzy-window-size parameter is 3. |
| – | Families contextually corrected—Families with the same fragment alignment coordinates and compatible UMIs are merged. |
| – | Duplex families—Families with close alignment coordinates and complementary UMIs are merged. |
When you specify a valid path for --umi-metrics-interval-file, DRAGEN outputs a separate set of on target UMI statistics that contains only families within the specified BED file.
If you need to analyze the extent to which the observed UMIs cover the full space of possible UMI sequences, the histogram of unique UMIs per fragment position metric may be helpful. It is a zero-based histogram, where the index indicates a count of unique UMIs at a particular fragment position and the value represents the number of positions with that count.
The following figures and table describe available UMI metrics.
Fig 1. Read pairs with duplex UMI
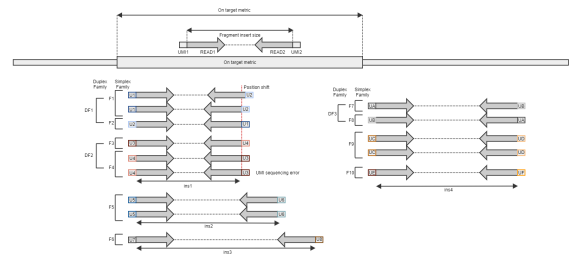
Fig 2. UMI error correction
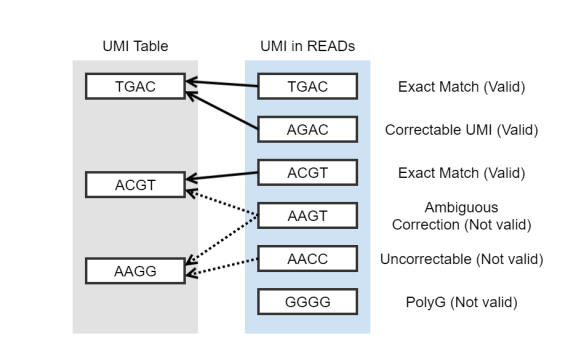
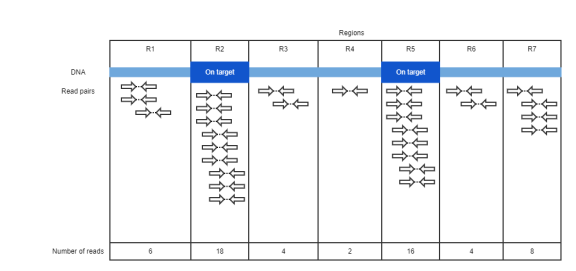
Fig 3. UMI collapsible regions
|
Metric |
Description |
Denominator of percentile |
Example |
|---|---|---|---|
|
Number of reads |
Total number of reads. |
N/A |
Fig 1. Read pairs with duplex UMI: 14 pairs of read X 2 = 28 reads |
|
Number of reads with valid or correctable UMIs |
Number of reads for which the UMIs could be corrected based on the lookup table. |
Number of reads |
Fig 2. UMI error correction: Valid UMI read count (Exact match+Correctable UMI) |
|
Number of reads in discarded families |
Number of reads in discarded families. Families are discarded when there are not enough raw reads to support the family. (family size less than "--umi-min-supporting-reads"). For "--umi-emit-multiplicity=duplex" option, simplex families will be discarded. |
Number of reads |
Fig 1. Read pairs with duplex UMI: Number of reads in Families discarded (See "Families discarded" for more detail) |
|
Reads with all-G UMIs filtered out |
Number of reads filtered out due to all-G in UMI sequence. |
Number of reads |
Number of reads in discarded families + Reads with all-G UMIs + Number of unpaired reads |
|
Reads filtered out |
Number of reads filtered out in total either for properties or in a discarded family. |
Number of reads |
Fig 2. UMI error correction: PolyG UMI read count |
|
Reads with uncorrectable UMIs |
Number of reads where the UMI could not be corrected. |
Number of reads |
Fig 2. UMI error correction: Uncorrectable + Ambiguous correction + PolyG |
|
Total number of families |
Number of simplex collapsed reads. |
N/A |
Read pairs with duplex UMI: F1~F10. |
|
Families contextually corrected |
Number of families that have some contextual correction. Contextual correction is based on other families at the same mapping location including UMI sequencing error and UMI jumping. |
Total number of families |
Fig 2. UMI error correction: Family count of correctable UMI |
|
Families shifted |
Number of families that have some shift correction. Shift correction merges families with fragment alignment coordinates up to the distance specified by the umi-fuzzy-window-size parameter. |
Total number of families |
Fig 1. Read pairs with duplex UMI: First read pair of DF1 (If shifted distance <= "umi-fuzzy-window-size") |
|
Families discarded |
Number of families filtered out by failing min supporting reads criteria or umi-emit type of simplex/duplex. |
Total number of families |
Fig 1. Read pairs with duplex UMI: Families discarded by min-support-reads + Families discarded by duplex/simplex (See below for detail) |
|
Families discarded by min-support-reads |
Number of families filtered out by failing min supporting reads criteria. |
Total number of families |
Fig 1. Read pairs with duplex UMI: Number of families size less than "umi-min-supporting-reads" option Size 1: F6, F10 Size 2: DF3, F5, F9 Size 3: DF1, DF2 |
|
Families discarded by duplex/simplex |
Number of families filtered out by failing umi-emit type of simplex/duplex. |
Total number of families |
Fig 1. Read pairs with duplex UMI: Number of simplex families (F5, F6, F9, F10) filtered. Note that simplex reads are only filtered if umi-emit-multiplicity=duplex (default: both) |
|
Families with ambiguous correction |
Number of families where the UMI cannot be corrected because more than one possible UMI corrections exists. |
Total number of families |
Fig 2. UMI error correction: Number of families of ambiguous correction UMI |
|
Duplex families |
Number of families that are merged as duplex (both strands). |
Consensus pairs emitted |
1. Read pairs with duplex UMI: DF1, DF2, DF3 |
|
Consensus pairs emitted |
Number of collapsed reads in output BAM. |
N/A |
Fig 1. Read pairs with duplex UMI: Depends on umi-emit-multiplicity=simplex/duplex/both, umi-min-supporting-reads=x simplex=F1~F10 (F2, F3, F6, F7, F8, F10 filtered if x>=2) duplex=DF1, DF2, DF3 both=DF1, DF2, DF3, F5, F6, F9, F10 (F6, F10 filtered if x>=2) |
|
Mean family depth |
Average number of read pairs per family. Filtered reads and families are excluded. |
N/A |
Fig 1. Read pairs with duplex UMI: Number of reads per family: DF1=3, DF2=3, DF3=2, F5=2, F6=1, F9=2, F10=1 Mean family depth = (3+3+2+2+1+2+1)/7 = 2 |
|
Histogram of num supporting fragments |
Number of families with zero raw reads, one raw read, two raw reads, three raw reads, etc. |
N/A |
Fig 1. Read pairs with duplex UMI: 0 reads: None 1 reads: F6, F10 = 2 (0 if umi-min-supporting-reads=2) 2 reads: DF3, F5, F9 = 3 3 reads: DF1, DF2 = 2 Histogram = {0|0|3|2} |
|
Number of collapsible regions |
Number of regions. |
N/A |
Fig 3. UMI collapsible regions: R1~R7 |
|
Min collapsible region size (num reads) |
Number of reads in the least populated region. |
N/A |
Fig 3. UMI collapsible regions: 2 reads (R4) |
|
Max collapsible region size (num reads) |
Number of reads in the most populated region. |
N/A |
Fig 3. UMI collapsible regions: 18 reads (R2) |
|
Mean collapsible region size (num reads) |
Average number of reads per region. |
N/A |
Fig 3. UMI collapsible regions: 8.3 |
|
Collapsible region size standard deviation |
Standard deviation of the number of reads per region. |
N/A |
Fig 3. UMI collapsible regions: 5.8 |
|
On target number of reads |
Number of reads that overlapped with UMI target interval --umi-metrics-interval-file. |
N/A |
Fig 1. Read pairs with duplex UMI, Fig 3. UMI collapsible regions: All On target metrics are same as corresponding metric but only considering fragments overlap with target intervals. i.e. DF3, F9, F10 in figure1 and R1, R3, R4, R6, R7 in figure3 are excluded from metric |
|
On target number of reads with valid or correctable UMIs |
Number of reads with a UMI that matched a UMI in the lookup table, including error allowance, and overlapped with UMI target interval. |
On target number of families |
|
|
On target number of reads in discarded families |
Number of reads in discarded families that overlapped with the UMI target interval. |
On target number of families |
|
|
On target duplex families |
Number of families that are merged as duplex among all the families that are overlapped with UMI target interval. |
On target number of families |
|
|
On target mean family depth |
Average number of reads per family that overlapped with UMI target interval. |
N/A |
|
|
On target families discarded |
Number of families that overlapped with UMI target interval filtered out by failing min supporting reads criteria or umi-emit type of simplex/duplex. |
On target number of families |
|
|
On target families discarded by min-support-reads |
Number of families that overlapped with UMI target interval filtered out by failing min supporting reads criteria. |
On target number of families |
|
|
On target families discarded by duplex/simplex |
Number of families that overlapped with UMI target interval filtered out by failing umi-emit type of simplex/duplex. |
On target number of families |
|
|
On target families with ambiguous correction |
Number of families that overlapped with UMI target interval where the UMI cannot be corrected because more than one possible UMI corrections exists. |
On target number of families |
|
|
Histogram of unique UMIs per fragment position |
Number of positions with zero UMI sequences, one UMI sequence, two UMI sequences, etc. |
N/A |
Fig 1. Read pairs with duplex UMI: 0 UMI sequence: None 1 UMI sequences: ins2 (F5), ins3 (F6) 2 UMI sequences: ins1 (DF1, DF2) 3 UMI sequences: ins4 (DF3, F9, F10) Histogram = {0|2|1|1} |
|
Total families in probability model estimation |
Total number of families used in estimation of UMI jumping rate and fragment size distribution used for probabilistic family merging. |
N/A |
|
|
Number of potential Jumping Families |
Total number of families that are potential UMI jumping candidates and the corresponding ratio. |
Total Families in Probability Model Estimation |
|
