Visualization and BigWig Files
To perform analysis on a known truth set, you can use the intermediate output files from the pipeline stages. These files can be parsed to aid in fine-tuning options.
All files have a structure similar to a BED file with an optional header line.
|
Option |
Description |
|---|---|
|
*.target.counts.gz |
Contains the number of read counts per target interval. This is the raw signal as extracted from the alignments of the BAM or CRAM file. The format is identical for both the case sample and any panel of normals samples. There is also a bigWig representation of a target.counts.diploid file, which is normalized to the normal ploidy level of 2 instead of raw counts. |
|
*.tn.tsv.gz |
The tangent normalized signal of the case sample, per target interval. This file contains the log-normalized copy ratio signal. A strong signal deviation from 0.0 indicates a potential for a CNV event. |
|
*.seg.called.merged |
Contains the segments produced from the segmentation algorithm. |
|
*.cnv.vcf.gz |
Output CNV VCF file that indicates events. |
To generate additional equivalent bigWig and gff files, set the --enable-cnv-tracks option to true. These files can be loaded into IGV along with other tracks that are available, such as RefSeq genes. Using these tracks alongside publicly available tracks allows for easier interpretation of calls. DRAGEN autogenerates IGV session XML file if tracks are generated by DRAGEN CNV. The *.cnv.igv_session.xml can be loaded directly into IGV for analysis.
The following IGV tracks are automatically populated in the output IGV session file:
|
Option |
Description |
|---|---|
|
.target.counts.bw |
BigWig representation of the target counts bins. Setting the track view in IGV to barchart or points is recommended. |
|
*.improper_pairs.bw |
BigWig representation of the improper pairs counts. Setting the track view in IGV to barchart is recommended. |
|
*.tn.bw |
BigWig representation of the tangent normalized signal. Setting the track view in IGV to points is recommended. |
|
*.seg.bw |
BigWig representation of the segments. Setting the track view in IGV to points is recommended. |
|
*.cnv.gff3 |
GFF3 representation of the CNV events. DEL events show as blue and DUP events show as red. Filtered events are a light gray. Selecting an event brings up a window for viewing annotation details. |
IGV Example
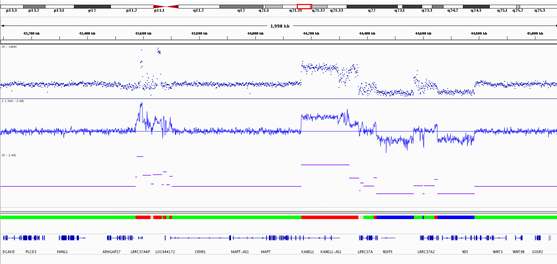
The IGV session XML file is prepopulated with track files generated by DRAGEN. The session file loads the reference genome that best matches the standard reference genomes in an IGV installation, by comparing the name of the --ref-dir specified on the command-line. Standard UCSC human reference genomes are autodetected, but any variations from the standard reference genomes might not be autodetected. To edit the genome detection, alter the genome attribute in the Session element to the reference genome you would like for analysis before loading into IGV. The reference identifier used by IV might differ from the actual name of the genome. The following is an example edited session file.
<?xml version="1.0" encoding="utf-8"?>
<Session genome="b37" hasGeneTrack="false" hasSequenceTrack="true" version="8">
<Resources>
<Resource path="example.cnv.gff3"/>
<Resource path="example.cnv.excluded_intervals.bed.gz"/>
<Resource path="example.target.counts.bw"/>
<Resource path="example.improper.pairs.bw"/>
<Resource path="example.tn.bw"/>
<Resource path="example.seg.bw"/>
</Resources>
<Panel height="500" width="1200" name="DataPanel">
...
</Panel>
</Session>
