ORA Format Specification
This document details the specification of the ora file format version 2.5.5. The ora format is a reference-based compression format for FASTQ files. It is a lossless compression format, designed for very fast compression/decompression and high compression ratio.
A patent is pending on the encoding methodology described in these specifications. The corresponding decoding methodology remains patent free.
Subdivision in Blocks
An ora file consists in a list of ora blocks. Each ora block contains up to 50000 reads and is self sufficient: all the information necessary for its decompression is contained within the block. There is therefore no overall file header containing additional information.
Reads in the ora file are ordered in the same way as the input FASTQ file format. If two paired FASTQ files are used as input, the reads are interleaved in the ora format (fragment 1 read 1, fragment 1 read 2, fragment 2 read1, fragment 2 read 2, ...).
This scheme has many advantages. First it makes possible the concatenation of compressed ora files: the concatenation of two lists of blocks is still a list of blocks and thus a valid ora file. Secondly it makes streaming possible, as soon as a single block is transferred its decompression can start. And lastly it makes parallel decompression easier, since each separate block can be handled independently from the rest.
Block Content
Figure 1 shows the content of an ora block. It starts with a header and is followed by a list of data sections. There are separate data sections for DNA sequences, read names, quality scores, sequence size, presence of ‘N’ characters in DNA sequences, and a crypto section possibly containing cryptographic keys. The quality scores are split into two different sections, one for sequences that contains at least one ‘N’ and the second one for reads that do not contain an ‘N’. This allows for different compression schemes for the two cases. The miscellaneous fields are for implementation dependent usage, as explained in a later section. Each data section can be of size 0 if unused. The block header contains in particular, among other metadata, the length in bytes of each of the data sections.
Figure 1: Content of an ora block.
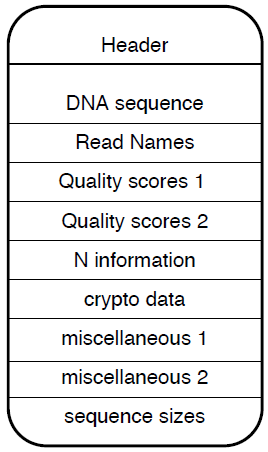
It starts with a header, followed by a list of data sections. The header of the block, contains, among other metadata, the length of each of the data sections. The ora block contains all the information necessary for its decompression.
In this document, the following data types are used:
|
Type |
Description |
|
uint8_t |
unsigned 8 bit integer |
|
uint16_t |
unsigned 16 bit integer |
|
uint32_t |
unsigned 32 bit integer |
|
uint64_t |
unsigned 64 bit integer |
|
int8_t |
signed 8 bit integer |
|
int16_t |
signed 16 bit integer |
|
int32_t |
signed 32 bit integer |
|
int64_t |
signed 64 bit integer |
The ora block header is formally described in table 2. All multi-bytes values are stored in little-endian fashion.
Each ora block must start with this header, with the first two bytes of the header being the magic value 0x7C49. Note that the header size is not fixed in the format, but must include the list of following required fields. As of version 2.5.5, the header is 121 bytes, but may be larger in future versions or include additional implementation-dependent custom data. A compliant decoder should get the header size from the second field of the header. The header size includes the size of the magic number and 'header size' field.
|
Field |
Description |
Type |
|
magic |
ora magic string 0x7C49 |
uint16_t |
|
l_header |
size of this header in bytes |
int32_t |
|
l_dna |
size of the DNA buffer in bytes |
uint32_t |
|
l_names |
size of the read names buffer in bytes |
uint32_t |
|
l_qualN |
size of the quality buffer 1 in bytes |
uint32_t |
|
l_qual |
size of the quality buffer 2 in bytes |
uint32_t |
|
l_size |
size of the sequence sizes buffer in bytes |
uint32_t |
|
l_N |
size of the N position info buffer in bytes |
uint32_t |
|
l_m1 |
size of the crypto buffer in bytes |
uint32_t |
|
l_m2 |
size of the miscellaneous buffer 1 in bytes |
uint32_t |
|
l_m3 |
size of the miscellaneous buffer 2 in bytes |
uint32_t |
|
flags |
bitwise flags |
uint32_t |
|
l_read |
read size |
int32_t |
|
n_reads |
number of reads stored in this block |
int32_t |
|
version |
ora version used to compress this block |
uint16_t |
|
b_id |
block ID |
uint64_t |
|
q_type |
type of quality scores |
uint8_t |
|
q4_1 |
Q4 quality symbol 1 |
uint8_t |
|
q4_2 |
Q4 quality symbol 2 |
uint8_t |
|
q4_3 |
Q4 quality symbol 3 |
uint8_t |
|
q4_4 |
Q4 quality symbol 4 |
uint8_t |
|
l_names_raw |
size of uncompressed read names |
uint32_t |
|
l_DNA_raw |
size of uncompressed DNA sequence |
uint32_t |
|
l_qual_raw |
size of uncompressed DNA quality scores 2 |
uint32_t |
|
l_qualN_raw |
size of uncompressed DNA quality scores 1 |
uint32_t |
|
l_qualTotal_raw |
size of uncompressed DNA quality scores |
uint32_t |
|
c_time |
epoch date of file creation |
uint64_t |
|
checksum_raw |
checksum of the uncompressed fastq data |
uint64_t |
|
checksum_ref |
checksum of the reference used for compression |
uint64_t |
|
checksum_comp |
checksum of this ora block |
uint64_t |
Additional field information :
| • | flags bitwise flags. List of flags is : |
|
Bit |
Description |
|
0x1 |
all reads in block have same size |
|
0x2 |
reads are interleaved, generated from pair of files |
|
0x4 |
read names are not stored |
|
0x8 |
read names are stored in tokenized mode (as described later) |
|
0x10 |
read names stored in fallback mode |
|
0x20 |
quality scores stored in fallback mode |
|
0x40 |
DNA sequence stored in fallback mode |
|
0x80 |
hybrid encryption mode activated |
|
0x100 |
unused |
|
0x200 |
unused |
|
0x400 |
unused |
|
0x800 |
unused |
|
0x1000 |
original input file name stored in misc1 buffer |
|
0x2000 |
input fastq file was gzipped |
| • | l_read |
This field contains the read size in base pairs when all reads in block have the same size (as indicated by flag 0x1). In that case the “sequence size” section of the block is empty. If reads in block have different sizes, their sizes are stored in the “sequence size” data section.
| • | n_reads |
Number of reads stored in this ora block.
| • | version |
Version is stored as 10000 major + 100 minor + release, e.g. 20505 for version 2.5.5.
| • | b_id |
Identifier of the block, mainly used for debugging purposes. It is not guaranteed to be in consecutive or increasing order.
| • | q_type |
Quality type of the fastq file. May either be 4 for files containing at most 4 different quality scores, or 40 for all other FASTQ files.
| • | Q4_1-4 |
When quality type is 4, the 4 fields Q4_1-4 contains the ascii characters of the 4 quality scores actually used in the FASTQ file.
| • | c_time |
Used to store the creation date of this ora block, as the value returned by the standard C library 'time' function. Note that the blocks of an ora file may have different creation dates (e.g. if a file was created as a concatenation of multiple files).
| • | checksum_raw |
Contains the checksum of the uncompressed data. It is computed with the xxhash function³.
| • | checksum_ref |
Contains the checksum of the reference genome file used for compression, computed with the xxhash function³.
| • | checksum_comp |
Contains the checksum of the block, computed with the xxhash function³. This checksum is computed from the block header and all the block data sections concatenated together, with the 'checksum_comp' field of the header set to 0.
Overall Description
ora is a reference based compressor. Reads are first mapped to a reference genome, and then encoded as a position in the genome plus a list of differences. The mapping algorithm is not a part of the specification, it is implementation dependent. Any mapper/aligner that provides basic information, e.g. such as read position on the reference genome, number and position of mismatches could be used. To be noted, the choice of read aligner plays an important role in the compression speed and compression ratio of an ora compliant compressor.
Sequences are encoded as a custom binary format defined below, which is then further compressed with an entropy coder.
DNA Sequence Binary Format
The sequence binary format is a list of read groups (i.e. an ora block contains a 'DNA sequence section' that itself consists in a list of read groups, with 'read group' defined below ). Each read group contains at most 8 reads. It starts with 4 bytes containing flag values for the following 8 reads, followed by 8 read records. The rationale for this grouping is to have easy to use byte aligned data: bit information per read is grouped with 8 reads to form a byte. Each read record contains the mapping data necessary to reconstruct the read from the reference at decompression.
The ora format currently does not use any kind of gapped information. Read alignments are therefore only limited to the following types:
| • | Perfect: alignment of the whole read, 0 mismatch. |
| • | Global: alignment of the whole read, at least 1 mismatch. |
| • | Local: alignment of a part of the read only. ( also known as clipped alignment ) |
| • | Unmapped : mapper was not able to map the read on the reference genome. |
One read group is defined as the following fields:
|
Field |
Description |
Type |
|
Perfect |
perfect mapping |
uint8_t |
|
Forward |
forward / reverse mapping |
uint8_t |
|
with N |
read contains N |
uint8_t |
|
pos_t |
|
uint8_t |
|
read records 0 to 7 |
|
list of read records |
Additional details:
• Perfect
One byte value. Bit i is set to 1 if i-th read of the group is perfectly aligned to the reference
(full-length mapping, 0 mismatch)¹.
• Forward
One byte value. Bit i is set to 1 if i-th read of the group is aligned to the forward strand, 0
if aligned in reverse.
• with N
One byte value. Bit i is set to 1 if i-th read of the group contains at least one ‘N’.
• pos_t
One byte value. Bit i is set to 1 if the field position of the i-th read of the group is encoded
with 16 bit mode as descirbed below.
• Read records 0 to 7
All read groups contains exactly 8 reads, except the last one of the ora block which contains 1 to 8 reads. The read group does not specifically store the number of reads encoded in the group, but this can be inferred from the total number of reads in this ora block stored in the block header.
¹with bit 0 denoting the most significant bit of the byte, and bit 7 the least significant bit.
The read record specification depends on the alignment type.
Perfect alignment:
If the alignment is perfect (as defined in the read group), then the read record is defined as:
|
Field |
Description |
Type |
|
position |
position of the alignment on the reference genome |
uint16_t / uint32_t |
The position can either be an absolute position in 32 bit on the reference genome (with all chromosomes concatenated to form one virtual single sequence), or a 16 bit incremental position from the previous read. Alignments on a larger than 232 bp reference are not supported in this format. The coding mode for the read is specified in the read group, as specified via the corresponding bit in the pos_t field.
Non-Perfect alignment:
If the alignment is not perfect, the read record will start with an 8-bit flag field containing, among other things, the type of the alignment. The remaining alignment types are global, local, raw, raw4. The 8-bit flag is of type flag8_t described below.
flag8_t:
The 8-bit flag8_t field is defined as (with bit 0 denoting the most significant bit of the byte, and bit 7 the least significant bit): 1with bit 0 denoting the most significant bit of the byte, and bit 7 the least significant bit.
|
Bit |
Description |
|
bit 0-4 |
number of mismatches |
|
bit 5 |
global alignment |
|
bit 6 |
raw alignment |
|
bit 7 |
raw4 alignment |
Bits 0 to 4 contain the number of mismatches in the alignment, it is therefore a value in 0-31. Bit 5 is set to 1 if the alignment is global, 0 otherwise. The distinction between the three other types isspecified by bits 6 and 7. A local alignment would have bits 5, 6 and 7 set to 0.
Global alignment:
Global alignment denotes that the whole read is aligned to the reference genome. Its read record is defined as:
|
Field |
Description |
Type |
|
flag8_t |
bitwise flags |
uint8_t |
|
position |
position of the alignment on the reference genome |
uint16_t / uint32_t |
|
mm_list |
list of mismatch entries |
variable size, list of mismatch_entry_t |
The number of entries in the mm_list (defined later) is equal to the number of mismatches in the read indicated in bits 0-4 of the flag8_t.
Local alignment:
Local alignment denotes that only a part of the read has been aligned to the reference genome. Its read record is defined as:
|
Field |
Description |
Type |
|
flag8_t |
bitwise flags |
uint8_t |
|
position |
position of the alignment on the reference genome² |
uint16_t / uint32_t |
|
mm_list |
list of mismatch entries |
variable size, list of mismatch_entry_t |
|
l_left |
length of unmapped left part |
uint8_t |
|
l_right |
length of unmapped right part |
uint8_t |
|
raw_nt |
raw unmapped nucleotides, encoded as 2 bit per nt |
variable size |
The raw_nt field contains the concatenation of unmapped left and right parts of the read. It is encoded as 2 bit per nucleotide, with values 0,1,2,3 coding respectively for A,C,G,T. The field size is rounded to the upper byte boundary, with the least significant bits of the last byte padded with 0. Figure 2 shows the encoding example of a local alignment.
²position in that case is the position on the reference genome of the first aligned bp of the read
Raw alignment:
In this mode the whole DNA sequence is encoded with 2 bits per nucleotide. It is mainly used for unmapped reads, but may be used in some other cases. 2 bit values 0,1,2,3 encode respectively for A,C,G,T.
|
Field |
Description |
Type |
|
flag8_t |
bitwise flags |
uint8_t |
|
raw_nt |
raw unmapped nucleotides, encoded as 2 bit per nt |
variable size |
The 'raw_nt' field size is rounded to the upper byte boundary, with the least significant bits of the last byte padded with 0.
Raw4 alignment:
This is the same as before, but using a 4 bits per nucleotide encoding mode. This encoding mode is used for unmapped reads that contains Ns. Values 0,1,2,3,4 encode respectively for A,C,G,T,N.
|
Field |
Description |
Type |
|
flag8_t |
bitwise flags |
uint8_t |
|
raw_nt |
raw unmapped nucleotides, encoded as 2 bit per nt |
variable size |
The 'raw_nt' field size is rounded to the upper byte boundary, with the least significant bits of the last byte padded with 0.
N list:
For reads that contain at least one ‘N’, as defined by the flag in the read group, then the read record contains at its end an additional field defining the list of position of ‘N’ in the sequence. It is defined as:
|
Field |
Description |
Type |
|
nb_N |
the number of N stored in the list |
uint8_t |
|
list_p |
list of N positions |
variable size, multiple of uint16_t |
The format imposes a maximum value of nb_N = 31. Reads that would require a larger number are instead encoded in the raw4 mode.
Each position in the list is a 0-based position in the read defined as a an incremental offset from the previous N position. For values below 215 the offset is coded as a 16 bit unsigned value. For offsets larger than or equal to 215 then multiple uint16_t are emitted, which are summed to produce the desired total offset.
mismatch_entry_t:
The mismatch entry type is a single byte defined as3 ³:
|
Bit |
Description |
|
bit 0-5 |
incremental position of the mismatch |
|
bit 6-7 |
alternate nucleotide |
The mismatch position is a 0-based position in the read defined as an incremental offset from the previous mismatch position, or from position 0 for the first mismatch of the list. The alternate nucleotide is a 2-bit value defining the nucleotide mismatch, with values 0,1,2,3 coding respectively for A,C,G,T.
³with bit 0 denoting the most significant bit of the byte, and bit 7 the least significant bit.
The capacity of several fields is deliberately limited to save space. The general idea is to cope only with the most frequent cases. Alignment data that would overflow some fields are handled differently, as follows.
The number of mismatches in an alignment is limited to 31 maximum as per the specification of flag8_t. If an alignment contains more than 31 mismatches, it cannot be encoded as a local or global alignment, and should be encoded as a raw alignment.
The incremental position of a mismatch is limited to 63 maximum as per the specification of the mismatch_entry_t. If a mismatch is more than 63 apart from the last mismatch, a “fake” mismatch will be inserted in between to allow for the encoding of all the mismatches. See figure 3 for an example.
Figure 2: Encoding example for a local alignment, showing the binary encoding of the flag8, position, mm_list, l_left, l_right and raw_nt fields
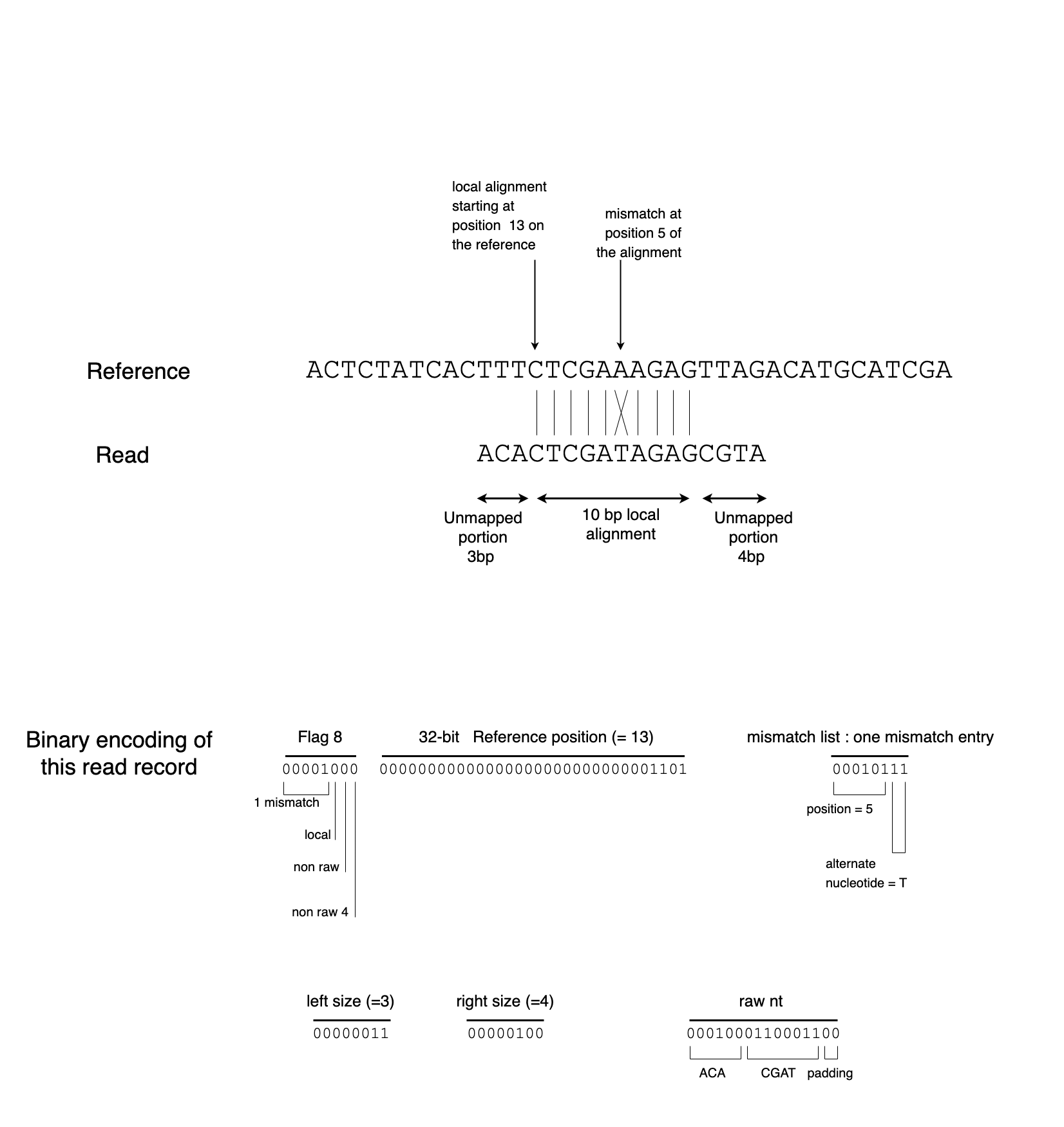
Figure 3: Encoding scheme of the mismatches.
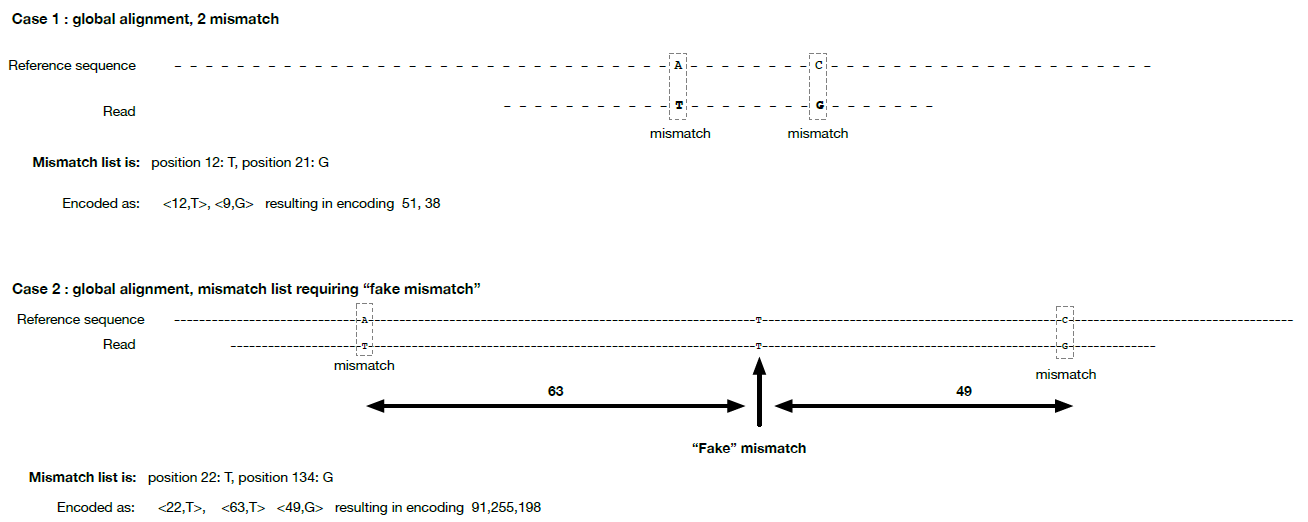
Each mismatch is encoded with a single byte, with first 6 bits being an offset from previous mismatch, and last 2 bits being the alternate nucleotide.
Case 2: If a mismatch is more than 63 apart from previous mismatch, one or several fake mismatches are inserted in between with alternate nucleotide (bits 6 and 7) equal to reference at that position.
Entropy coder for the DNA sequence data section
The DNA sequence data section consisting of the list of read groups as defined previously is then compressed with the zstd library⁴. Compression level of the zstd library is implementation dependent.
Fallback mode
DNA sections may alternatively be compressed in a Fallback mode as specified in the bit 0x40 of the flags of the block header. In this mode, all DNA sequences are concatenated together and compressed with zstd library, with an implementation dependent level.
Overall description
Quality scores are compressed in a completely lossless way. The overall scheme is to apply some transformations, then use a context model followed by a range encoder. The transformations and context models used are adapted to different kind of quality encodings. ora distinguishes between quality scores generated by sequencers that generate quality on 4 different levels, and other type of sequencers that generate more levels. These two types are respectively called Q4 and Q40 in the ora format.
Among a type, we further distinguish between reads that contain at least one ‘N’ and other reads. The idea is to further exploit specificities of the encoding: for Q4 qualities, the 0 quality value is usually reserved for ‘N’ in the sequence. Therefore if the read does not contain an ‘N’ then its quality score string only contains 3 different levels.
Q40 Quality Scores
This quality mode applies to quality strings that contains a range of at most 64 different quality values.
For Q40 quality scores, the first transformation applied during compression is to replace some frequent trimers by a single 1 byte code.
At compression, the 190 most abundant trimers (words of 3 quality values, i.e a word Q₁Q₂Q₃) are computed from all the overlapping trimers of quality strings in the ora block. The data section starts with a table of size 190 x 4 Bytes containing the trimer value of each of the 190 most abundant trimer used. The trimer value is defined as (Q1–Qlowest)+(Q2–Qlowest)x64+(Q3–Qlowest)x64². Each trimer value is also mapped to trimer code in the range [64-254], as ordered in the table. Qlowest is the lowest quality value observed in the block, and is stored right after the 190-table as a single byte. Each quality value is transformed as follow :
| • | If quality value is among an abundant 190 trimer: |
Q₁Q₂Q₃←trimercode[Q₁Q₂Q₃]
| • | else |
Q←Q–Qlowest
After transformation, the string of quality values is a list of bytes representing either a single quality value ( if byte value in the range 0-63), or a word of 3 consecutive qualities (byte value in the range 64-254).
The beginning of the quality data section is in that case:
|
Field |
Description |
Type |
|
190-table |
trimer codes of the 190 most frequent trimers |
190 x uint32_t |
|
lowest_val |
lowest quality value in the block |
uint8_t |
The string of transformed quality values are then encoded with a range encoder. The context model used in this cases is an 8 bit context consisting in the last 8 previous 8 bit in the string. The range encoder used is from PPmd².
Q4 quality scores
The Q4 type applies to quality strings that contain at most 4 different quality values. The quality strings of each read is further divided into two groups, one group for strings that do not contain the 0 value (and hence only have 3 different quality values), and the rest.
The four different quality values originally in ASCII range are converted to four values in the 0-3 range. The block header contains in the q4* fields th association between the 0-3 range and the four corresponding ASCII values.
This is the subgroup with quality values in the range 0-3 included . In practice it usually corresponds to a small fraction of total data, i.e. only reads that contains N.
Binary format
Quality values are first binary encoded on 2 bit per quality value.
Entropy coder
The data string is then compressed with the zstd library⁴. Compression level used is implementation dependent.
This is the subgroup with quality values in the range 1-3.
Binary format
Five consecutive quality values q₁q₂q₃q₄q₅ in the range 1-3 are encoded on a single byte with the formula :
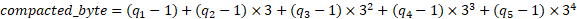
The compacted_byte is a number with 5 digits in base 3. This corresponds to a 1.6 bit encoding per quality value.
Entropy coder
The compacted 1.6 bit / quality value data stream is further compressed with a range encoder. The range encoder used is from PPmd². The context model consists in a 4-bit value representing the number of highest value quality scores in the preceding 30 quality scores.
Fallback mode
Quality scores may alternatively be compressed in a Fallback mode as specified in the 0x20 bit flag of the block header. In this mode, all quality scores of the data section are concatenated together and compressed with zstd library, with an implementation dependent level.
Overall description
Read names are tokenized based on standard C lib isdigit function. The read name is split into substrings containing only digits or only non digit characters. Read names of an ora block are encoded in a tokenized fashion if all read names in the block have the same number of tokens. They are otherwise encoded in the fallback mode described below.
Then i-th token of each read name are concatenated together into a token set. Digit tokens are binarized and then encoded as incremental value from the token of previous read name. Each token set is then compressed using zstd library⁴.
Binary format
The read names section is defined with the following fields :
|
Field |
Description |
Type |
|
nb_tokens |
number of token sets |
uint32_t |
|
token_type_list |
list of types of each token set |
uint8_t |
|
token_set_size_list |
list of size of each token set |
uint64_t |
|
token_sets_list |
list of all token sets |
variable size |
| • | nb_tokens The number of token in read names. This means there will be nb_tokens token sets stored in the token_sets_list. token_type_list Type of each token set. Types are encoded as a value in the 0-5 range corresponding to types : string,numeric,numeric_incr, numeric_incr16, numeric_incr8, numeric_ incr32 |
| • | token_set_size_list The size of each token set in bytes, encoded on 64 bit unsigned integer. |
| • | token_sets_list The list of all token sets. Each token set is compressed according to its type. String tokens consists in the concatenation of this token for all read names of the group then compressed with zstd library. numeric_incr tokens are encoded as differential value from token in the preceding read, and encoded with 8, 16 or 32 bit unsigned integer. The value for the first read is encoded as a 64 bit signed integer. |
Fallback mode
If read names in the block do not all have the same number of tokens, then read names are compressed in this fallback mode. In that case read names are concatenated together, with each read name delimited by a null byte. All printable ASCII characters are allowed in read names. It is then compressed with zstd library, with level implementation dependent. The mode used to compress read names is recorded in the ora block header, with bit flag 0x10.
This section encodes one boolean value per read, true if the read contains at least one N. The information is redundant from the flag present in the read group but is duplicated here for practicality reasons. Its size is negligible. It is encoded as a bit array, then compressed with the Zstd library, with implementation dependent level.
This data section encodes the list of DNA sequence sizes stored in the DNA sequence data section in the case they are not all the same in this block. If they are all the same then this section is empty and read size is stored in the block header once.
The sizes are encoded by group of 8 size values called a sequence size group. Each group starts with the flags:
|
Field |
Description |
Type |
|
same_flag |
same size as previous read |
uint8_t |
|
ize_flags |
defines the encoding type of the size |
uint8_t |
| • | same_flag |
One byte value. Bit i (with bit 0 the most significant bit) is set to 1 if i-th size in the group is equal to the previous value.
| • | size_flag |
One byte value. Bit i is set to 1 if i-th size of the group is encoded on a single byte. After the two flags values, each size is encoded according to its type:
| • | same size In the case the same flag is set, then no size is stored, it equals the previous one. |
| • | size < 256 If size_flag is set to 1, then the read size is encoded as a single byte value. |
| • | size > 256 if both the same_flag and size_flag are set to 0 then the read size is encoded as a list of 16 bit unsigned integers. The size S is decomposed with the euclidian division S = QK + R, with K = 32768, and then stored as a list of Q+ 1 16 bit unsigned integers: Q times value 32768 and one time value R. This is a kind of prefix code tuned for small sequences. |
The data section is then further compressed with zstd library, with implementation-dependent level.
An ora block holding zero sequence (as specified in the block header n_reads variable) may be used to store additional metadata, by writing custom information in the “Miscellaneous 1” or “Miscellaneous 2” data sections of the block. Information stored in these data sections is implementation dependent.
As an example, this may be useful to hold file-level statistics with a zero-sequence block at the end of the file. A format compliant decoder may just ignore ora blocks holding zero sequence and skip them.
Overview
The ora format optionally supports encryption of the file with an hybrid scheme. Data sections are encrypted using a symmetric cipher, whose encryption key is stored in the file after encryption by an asymmetric cipher.
From the user perspective, the file is encrypted with an asymmetric scheme: a public key is used for encoding the file, and the associated private key is required to decipher and read the file. Internally, a new key K is randomly generated for the symmetric cipher for each new ora block, and this key is encrypted with the asymmetric cipher with the user provided public key and stored in the block. Data sections of the block are encrypted with the randomly generated key K. When reading the file, the key for the symmetric cipher is deciphered with the user provided private key, and use to decipher the data section with the symmetric cipher.
This scheme allows for the convenience of an asymmetric scheme for the user, while retaining the high speed of a symmetric cipher for the encryption of the file. The symmetric cipher used is AES with a 128 bit key, and the asymmetric cipher is RSA with 4096 bit key. Implementations of RSA and AES are those provided by the CryptoPP library¹.
Cipher keys
When the file is encrypted, the “crypto data” section hold the following data :
|
Field |
Description |
Type |
|
RSA-digest |
fingerprint of public RSA key |
32 bytes |
|
magic |
ora magic string 0x7C49 |
uint16_t |
|
aes |
AES key |
uint128_ |
Fields “magic” and “aes” are stored encrypted with RSA with the user-provided public key. A 32 bytes RSA-digest of the RSA public key used is stored in the file for user convenience. The ora magic string encrypted with RSA may be used as a quick test when trying to decrypt an encrypted ora file to check if the user provided private key is the correct one.
Encryption
When encryption is activated all data sections are individually encrypted with AES with a 128 bit key. The header of each ora block is not encrypted and contains in particular a flag specifying if the block is encrypted or not. Each block is encrypted with AES in CTR mode with a 128 bit initialization vector. Each data section has its own randomly generated initialization vector that is stored at the end of the data section.
| 1. | CryptoPP library: https://github.com/weidai11/cryptopp. Accessed: 2021-03-1. |
| 2. | PPmd: http://compression.ru/sh/ppmy_3c.rar Accessed: 2020-04-3. |
| 3. | xxhash function: https://github.com/Cyan4973/xxHash. Accessed: 2020-04-3. |
| 4. | zstd library: https://github.com/facebook/zstd/. Accessed: 2020-04-3. |
