Attaching Files Located Outside the Default File Storage Repository
Compatibility: API version 2 revision 14 - 24
In Next Generation Sequencing (NGS), a large amount of data is generated in a single run. Since the volume of data is so large, it makes sense to link to data files as they exist on the network, rather than copying files to the Clarity LIMS file server.
This example shows how you can use the files resource to associate a process output result file with a file on the network using the HTTP protocol.
| • | You have added samples to the system. |
| • | You have defined a Next Gen Sequencing process that takes analyte (derived sample) inputs and creates a single result file output for each input. |
| • | You have defined a flow cell container with 8 rows and 1 column. Both the rows and columns are numbers and start at 1. |
| • | You have added samples to a flow cell and have run the flow cell through your Next Gen Sequencing process. |
| • | You have defined and set up an HTTP file store in the LIMS configuration file, in accordance with the instructions in Setup, below. |
| • | You have added the appropriate directory structure and files to be linked to your HTTP file store.For this example, you will need a directory with the same name as the flow cell on which you run your process. |
| – | You will require a subdirectory for each lane named by lane number eg "1" containing the file you want to link. |
| – | The name of the file must be easily programmatically-determined. For example s_1_export.txt for lane number 1. |
This example assumes you are using the standard Non-Production Scripting Sandbox Server, which uses Apache to serve files with HTTP.
If you plan to use an alternative file storage configuration, contact the IT adminstrator of the LIMS server.
The adminstrator will use the omxprops-ConfigTool.jar configuration tool, described in the Non-Production Scripting Sandbox Server - IT Admin Guide document.
The omxprops-ConfigTool.jar tool is located at:
/opt/gls/clarity/tools/propertytool/
For more information see Remote HTTP Filestore Setup.
After running a Next Gen Sequencing process on a full flow cell, in the Operations Interface, the Input/Output Explorer for the process run shows the relationship between the inputs in the flow cell and the result file output placeholders.
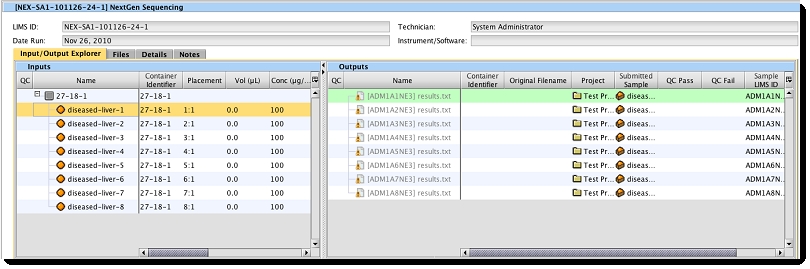
Associating a file that resides on a server that is accessible via the HTTP protocol requires the following steps:
| 1. | Matching up the file with the correct file placeholder. |
| 2. | Constructing the XML to POST to the files resource. |
| 3. | A POST to the files resource, which associates the file and creates the link between the artifact and the file. |
Step 1. Match the file with the correct file placeholder
The following code snippet shows how, by starting with only the name of the flow cell, we can obtain the API resource for the flow cell container and the Next Gen Sequencing process run.
Since the location of all the files we want to link is known, the combination of the flow cell and Next Gen Sequencing process contains all the additional information required to link each file to the correct file placeholder, so that the results will be associated with the correct sample.
| 1. | To begin, get a containers list filtering by container name, where the name is equal to the name of the directory. Your container name must be unique in the system as only one result is expected by this script. |
def containers = GLSRestApiUtils.httpGET("${baseURI}containers?name=${containerName}", username, password) println(containers)
| 2. | Now get the full XML for the container - the container list only contains the URI and LIMS ID of the container and we require all the placements as well. |
def container = GLSRestApiUtils.httpGET(containers.'container'[0].@uri, username, password)
| 3. | Using the LIMS ID of one of the placements in the container, use the API to find the processes that have used it as an input. For this example, we expect that only one process has been run on the flow cell; if that is not the case, the first one returned will be used. |
def processes = GLSRestApiUtils.httpGET("${baseURI}processes?inputartifactlimsid=${container.'placement'[0].@limsid}", username, password)
| 4. | Just as for the container case, the list resource just gives us the URI and LIMS ID of the process, but we need the complete XML with input-output-map. We need to make another call to the API to retrieve the complete process. |
def process = GLSRestApiUtils.httpGET(processes.'process'[0].@uri, username, password)
The code in this example includes some helper closures and a persistent single HTTP connection. The persistent connection helps improve the performance of the script.
The location of the file to be linked is constructed for each POST, based on the directory structure. For each input-output-map element in the process it is stored in the FileURI variable. The value provided here is used in both the <content-location> and <original-location> elements in the file node that will be posted.
String placement = container.placement.find{ it['@limsid'] == inputlimsid }.value[0].text() String laneNum = placement.split(':')[0] String fileURI = "${illuminaRunDirectoryURI}/${laneNum}/s_${laneNum}_export.txt"
The location of the result file artifact is stored in the variable outputURI. It is obtained from the <output> element in the i nput-output-map:
String outputURI = it.output[0].@uri.split("\\?")[0]
Step 2. Construct the XML to POST to the files resource
The first requirement is to create an XML resource for the file you want to submit to the system.
As in the Attaching a File to a Process Placeholder with REST example, you will create the resource using StreamingMarkupBuilder, which you will use in a POST to the files resource.
When you construct the individual file XML, you must specify the content-location, attach-to, and original-location child nodes.
In this example we are posting a file link to a file in a HTTP file storage location, so the content-locatio n and original-location are the same.
def fileBuilder = new StreamingMarkupBuilder() fileBuilder.encoding = "UTF-8" def fileXML = fileBuilder.bind { mkp.xmlDeclaration() mkp.declareNamespace(file: 'http://genologics.com/ri/file') 'file:file'{ 'content-location'(fileURI) 'attached-to'(outputURI) 'original-location'(fileURI) } }
Step 3. POST to the files resource
The POST requires an XML node as input. Therefore, you must convert fileDoc from a writable closure to an XML node using GLSGeneusRestApiUtils.xmlStringToNode.
You can then submit the new file resource to the API via a POST.
// Convert the fileXML to a node and link the file by posting the node to the files resource in the API Node fileNode = new XmlParser().parseText(fileXML.toString()) GLSRestApiUtils.httpPOST(fileNode, "${baseURI}files", username, password)
Below is an example of the XML for FileNode, which is used in the POST:
<?xml version='1.0' encoding='UTF-8'?><file:file xmlns:file='http://genologics.com/ri/file'> <content-location>http://Root of your http file store/illumina/27-18-1/1/s_1_export.txt</content-location> <attached-to>http://yourIPaddress/api/v2/artifacts/ADM1A1NE3</attached-to> <original-location>http://Root of your http file store/illumina/27-18-1/1/s_1_export.txt</original-location></file:file>
The POST command does the following:
| • | POSTs the XML constructed by StreamingMarkupBuilder to the files resource. |
| • | Adds a link from the files list resource to the new file. |
| • | After the POST executes, a link to the new file resource is added to the ResultFile artifact resource – because it was specified in the attached-to field of the new file resource. |
Below is the XML for the ResultFile artifact after the POST, which contains a link to the new file resource on the last line:
<art:artifact uri="http://yourIPaddress/api/v2/artifacts/ADM1A1NE3?state=48" limsid="ADM1A1NE3"> <name>results.txt</name> <type>ResultFile</type> <output-type>ResultFile</output-type> <parent-process uri="http://yourIPaddress/api/v2/processes/NEX-SA1-101126-24-1" limsid="NEX-SA1-101126-24-1"/> <qc-flag>UNKNOWN</qc-flag> <sample uri="http://yourIPaddress/api/v2/samples/ADM1A1" limsid="ADM1A1"/> <file:file limsid="ADM1A1NE3-40-112" uri="http://yourIPaddress/api/v2/files/ADM1A1NE3-40-112"/></art:artifact>
The file resource available from the API is slightly altered from the XML submitted to the POST. The file is assigned a LIMS ID and a URI.
Below is the XML resource for the file that is available from the API:
<file:file uri="http://yourIPaddress/api/v2/files/ADM1A1NE3-40-112" limsid="ADM1A1NE3-40-112"> <attached-to>http://yourIPaddress/api/v2/artifacts/ADM1A1NE3</attached-to> <content-location>http://Root of your http file store/illumina/27-18-1/1/s_1_export.txt</content-location> <original-location>http://Root of your http file store/illumina/27-18-1/1/s_1_export.txt</original-location> <is-published>false</is-published></file:file>
|
The POST to the files resource associates a physical file to the ResultFile artifact. In the user interface, this POST changes the file icon. |
Before POSTing to the files resource, you must make sure that the file exists in the location referenced by the content-location element. If the file does not exist in this location, the POST will fail. |
The value returned from the POST is stored in the returnNode variable.
| • | If the command was successful, the XML as shown above is returned. |
| • | If the command was unsuccessful, an XML document explaining the error is returned. For example, if a file was already attached to the result file located at artifactURI, the document shown below is stored in returnNode, as returned from the POST. In this case, the file that is already attached to the artifact must first be removed via the file resource DELETE method. |
<exc:exception xmlns:exc="http://genologics.com/ri/exception"> <message>Removing file in this manner not permitted</message></exc:exception>
After a successful POST HTTP command, in the Operations Interface, the process summary view's Input/Output Explorer shows all the attached files.
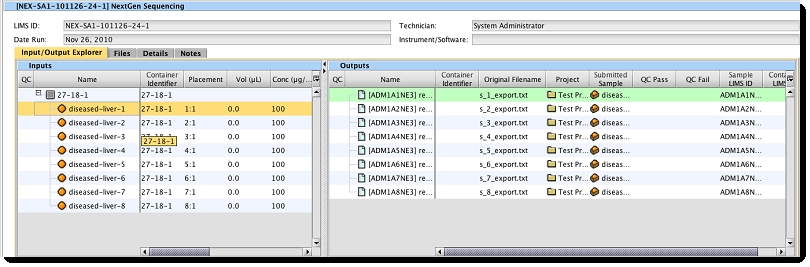
Attachments

