Illumina HiSeq/GAIIx Integration v5.5 / v5.6 User and Configuration Guide
Compatibility: See Illumina HiSeq/GAIIx Release Notes (for Clarity LIMS 4.0 or later).
Contents
The Illumina HiSeq/GAIIx Integration Package v5.5 supports Illumina HiSeq 1000, 1500, 2000, 2500, GAII(x), and HiScanSQ instruments.
The integration provides automated generation of a sample sheet file for use with the HiSeq or GAII(x) sequencing control software (HCS/SCS) and with bcl2fastq v2.15 or CASAVA v1.8.2 analysis software, and allows for automated tracking of an Illumina sequencing run in BaseSpace Clarity LIMS. Tracking includes instrument run status, automated reporting, and capturing and parsing of run statistics.
| • | This page describes the integration between BaseSpace Clarity LIMS and the IlluminaHiSeq/GAII(x) system. It includes information about protocols and automations, script parameters and usage, configuration options, installed components, and rules and constraints. |
| • | For instructions on validating and troubleshooting the Illumina HiSeq/GAIIx Integration Package v5.5, see the Illumina HiSeq/GAIIx Integration v5.5 / v5.6 Validation and Troubleshooting Guide. |
| • | For overview information about how the Illumina – BaseSpace Clarity LIMS interface works, and which events are monitored, see How the Illumina Instrument and LIMS Software Interact. |
This Illumina HiSeq/GAIIx Integration includes the following:
| • | Preconfigured TruSeq Exome Enrichment 5.0, Illumina SBS (HiSeq GAIIx) 5.0, and HiSeq/GAIIx Validation protocols |
| • | Automatic generation of sample sheet files for use with bcl2fastq v2.15 or CASAVA v1.8.2 analysis software |
| • | Automated tracking in BaseSpace Clarity LIMS of the progress and metrics of an Illumina sequencing run |
| • | Per-instrument tracking of sequencing runs |
| • | Ability to directly monitor run status (cycle number) across multiple instruments from within BaseSpace Clarity LIMS |
| • | Automated tracking of Real Time Analysis (RTA) run directory location and other run specific information |
| • | Automated tracking of sequencing run parameters |
| • | Automatic generation of a report, which includes the current run’s statistics and a comparison to the last five runs for any statistics that are one standard deviation off |
| • | Integration with Illumina's Bcl to FASTQ conversion and demultiplexing tool for CASAVA |
Information on these features is provided in the following sections.
The following out-of-the-box protocols are included in the Illumina HiSeq/GAIIx Integration Package v5.5:
| • | TruSeq Exome Enrichment 5.0 |
| • | Illumina SBS (HiSeq GAIIx) 5.0 |
| • | HiSeq/GAIIx Validation |
Figures 1 - 3 show these protocols as they display to the lab scientist working in Platform SW BaseSpace Clarity LIMS Lab View.
TruSeq Exome Enrichment 5.0 Protocol
Figure 1: Platform SW BaseSpace Clarity LIMS Lab View showing out-of-the-box TruSeq Exome Enrichment 5.0 protocol and steps
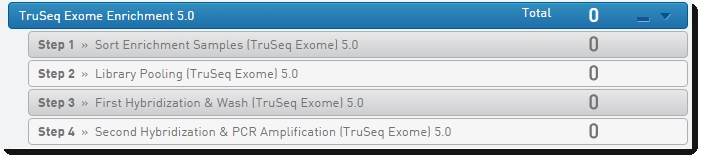
Illumina SBS (HiSeq GAIIx) 5.0 Protocol
Figure 2: Platform SW BaseSpace Clarity LIMS Lab View showing out-of-the-box Illumina SBS (HiSeq GAIIx) 5.0 protocol and steps
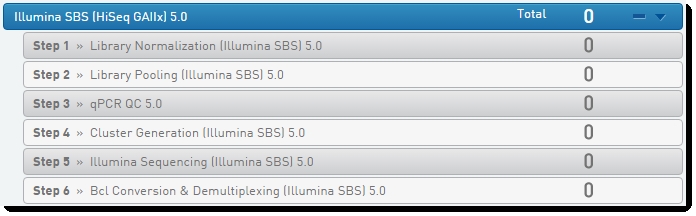
HiSeq/GAIIx Validation Protocol
Figure 3: BaseSpace Clarity LIMS Lab View showing HiSeq/GAIIx Validation protocol and step
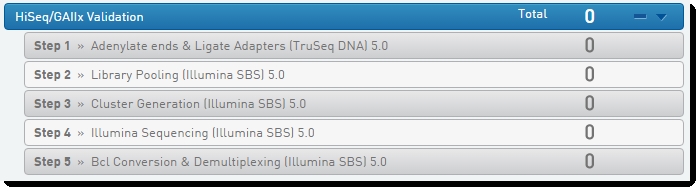
For instructions on using the HiSeq/GAIIx Validation workflow to validate automated sample sheet generation, see the Illumina HiSeq/GAIIx Integration v5.5 / v5.6 Validation and Troubleshooting Guide.
Automations
Table 1 lists the automations included in this integration, and the protocol steps on which they are configured. Additional details are provided in the sections following the table.
Note that the TruSeq Exome Enrichment 5.0 protocol does not include any automation.
Table 1: Illumina HiSeq/GAIIx Integration out-of-the-box protocols and automations
|
Protocol |
Automation and step |
|||||||||
|
Illumina SBS (HiSeq GAIIx) 5.0 |
|
|||||||||
|
Illumina SBS (HiSeq GAIIx) 5.0 HiSeq/GAIIx Validation |
One of these automations is disabled upon installation of the integration. See Configuring Sample Sheet Generation. |
|||||||||
Depending on the requirements of the system, the BCL Conversion and Demultiplexing (Illumina SBS) 5.0 step may be removed from the protocol. This is typically the case when the system is configured to use the bcl2fastq2 sample sheet for downstream analysis. See Configuring Sample Sheet Generation. |
The following sections describe how these automations work, and how the lab scientist interacts with them in the lab.
For more information on automations, including how to configure them, see the following topics in the LIMS Documentation.
LIMS v4.1 and v4.2 Documentation:
| • | Configuring Protocol Steps (Protocols section) |
| • | Passing Parameters via EPP (Integrations via EPP section) |
LIMS v5 Documentation:
| • | Configuring Protocol Steps (Protocols section) |
| • | Passing Files and Tokens to Third-Party Programs (Automations section) |
As of Platform SW BaseSpace Clarity LIMS v5.0, the term user-defined field (UDF) has been replaced with custom field in the user interface (the API resource is still called udf).
There are two types of custom fields:
| • | Master step fields - configured on master steps. These fields only apply to the master step on which they are configured, and the steps derived from those master steps. |
| • | Global fields - configured on entities such as submitted sample, derived sample, measurement, etc. These fields apply to the whole Clarity LIMS system. |
Library Normalization File Generation
See Normalization Buffer Volumes.
On the Record Details screen of the Library Normalization (Illumina SBS) 5.0 step:
| 1. | Enter values for each of the following: |
| • | Library volume (uL) transferred to destination plate |
| • | Normalized conc (nM) |
| • | Maximum destination container volume (uL) |
| • | Any custom fields/UDFs |
| 2. | Click Create Normalization CSV. |
Figure 4: Record Details screen of Library Normalization (Illumina SBS) 5.0 step
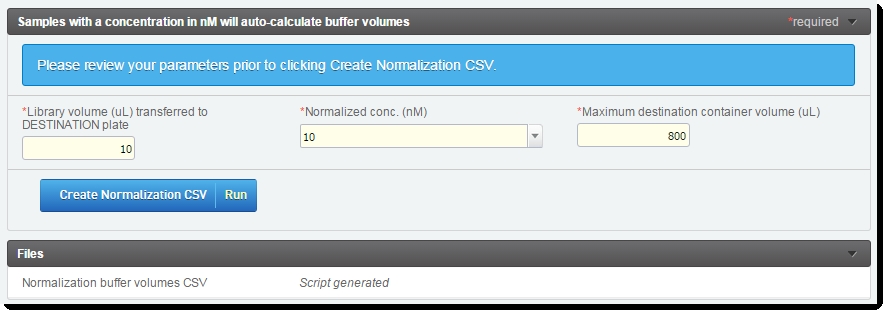
| • | Once the script has completed, the Normalization buffer volumes CSV file is generated and attached to the step. |
The assignQC script compares the values entered into the Sample Details table by the lab scientist with the criteria values set on the Record Details screen, and uses this information to set appropriate pass or fail QC flags on the output samples.
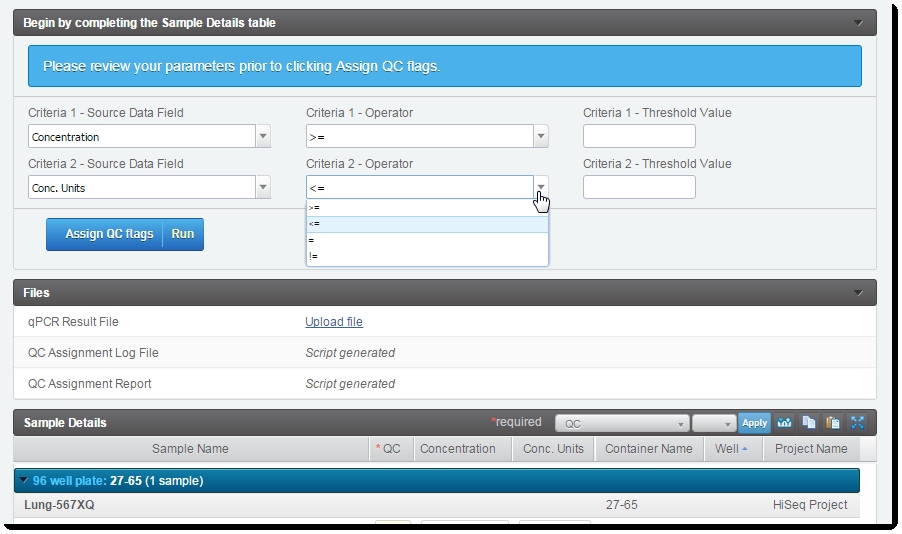
On the Record Details screen of the qPCR QC 5.0 step:
| 1. | Upload the qPCR Result File to the placeholder and manually complete the Sample Details table. |
| 2. | Set the desired Criteria Source Data Fields, Criteria Operators, and Criteria Threshold Values. |
| 3. | Click Assign QC flags. |
Figure 5: Record Details screen of qPCR QC 5.0 step
| 4. | Once the script has completed: |
| • | QC flags are set on the output samples |
| • | The QC Assignment Log File and QC Assignment Report are generated and attached to the step |
The integration allows for automatic generation of two sample sheets:
| • | One sample sheet designed to be used with the instrument control software |
| • | One sample sheet designed to be used with CASAVA v1.8.2 or bcl2fastq v2.15 sequencing analysis software. One of these analysis options is disabled when the system is initially configured – see Configuring Sample Sheet Generation. |
By default, one instrument sample sheet and one analysis sample sheet are created for the flow cell loaded during the Cluster Generation (Illumina SBS) 5.0 step.
If required, you may configure the step to generate multiple analysis sample sheets, and to generate sample sheets for multiple flow cells.
Performing analysis with CASAVA or bcl2fastq2 software is optional. If you prefer to use a different analysis pipeline, you will need to create your own sample sheet. You can use Illumina Experiment Manager to do this.
Quick Links:
| • | Guided sample sheet creation and setup |
| • | Illumina Experiment Manager User Guide |
| • | Illumina Experiment Manager Support |
Table 2: Sample sheet files and placeholders generated by Cluster Generation (Illumina SBS) 5.0 step
|
Item |
Description |
||||||
|
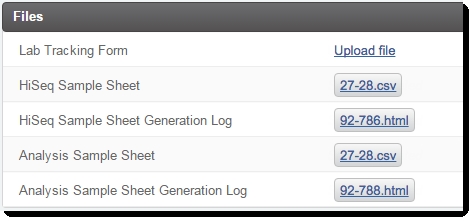
Lab Tracking Form placeholder |
This placeholder in the LIMS allows the user to manually attach a lab-specific tracking form to the step. |
||||||
|
HiSeq Sample Sheet (CASAVA format) |
This CSV file is automatically generated by the LIMS for use with the HiSeq/GAII(x) instrument.
|
||||||
|
Sample Sheet Generation Log |
Automatically generated by the LIMS, this log file captures any errors that the LIMS may encounter when generating the HiSeq/GAIIx sample sheet. |
||||||
|
Analysis Sample Sheet (for use with CASAVA or bcl2fastq2) |
This CSV file is automatically generated by the LIMS for use in CASAVA or bcl2fastq2 post-sequencing analysis.
|
||||||
|
Analysis Sample Sheet Generation Log |
Automatically generated by the LIMS, this log file captures any errors that the LIMS may encounter when generating the analysis sample sheet. |
| 1. | Sample sheet generation is configured on the Record Details screen of the Cluster Generation (Illumina SBS) 5.0 step – this is when the samples are loaded onto the flow cell that will be placed in the instrument. |
Figure 6 and Figure 7 show the three automations configured on the step, in LIMS v4.x and LIMS v5.x.
Figure 6: Cluster Generation (Illumina SBS) 5.0 automation configuration - LIMS v4.x
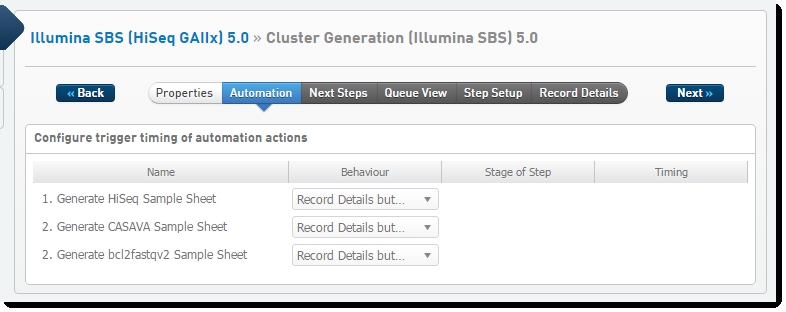
Figure 7: Cluster Generation I (Illumina SBS) 5.0 automation configuration - LIMS v5.x
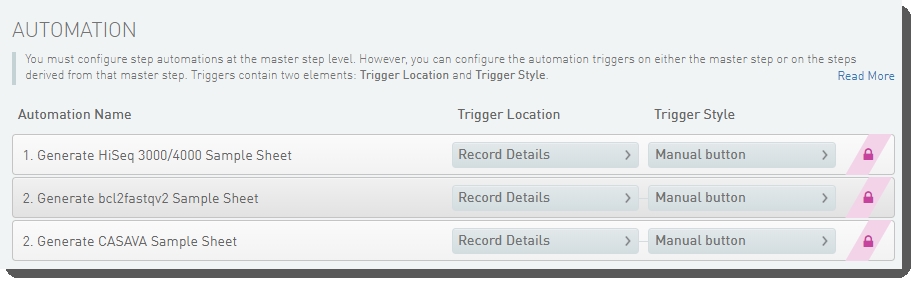
| 2. | Each of these automations invokes a script that generates the sample sheet – a CASAVA or bcl2fastq2 format comma-separated text file containing the required columns. |
| 3. | If the CASAVA format analysis sample sheet is used, once the sequencing run is in progress on the instrument and run data capture begins, the sample sheet for the current run is copied to the following directory for use with Illumina’s Bcl to FASTQ conversion and demultiplexing tool for CASAVA: |
<run data directory>/Data/Intensities/BaseCalls/
See the CASAVA 1.8.2 Sample Sheet Generation and bcl2fastq2 Sample Sheet Generation articles for detailed information on:
| • | Sample sheet generation script parameters and usage |
| • | Sample sheet data and configuration options |
| • | Sample sheet rules and constraints |
| 1. | On the Placement screen of the Cluster Generation (Illumina SBS) 5.0 step: |
| • | Scan the flow cell ID (FCID) to populate the Container Name field. |
| • | Place the samples and proceed to Record Details screen. |
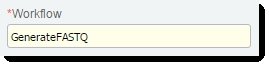
| 2. | On the Record Details screen of the Cluster Generation (Illumina SBS) 5.0 step: |
| • | Note that the value of the Workflow field is already set to GenerateFASTQ. |
Figure 8: Workflow field of Cluster Generation step
| • | In the Cluster Generation Kit field, select the desired kit from the drop-down list. |
| • | In the Cluster Generation Workflow field, select Paired-end or Single End Readfrom the drop-down list. |
Figure 9: Cluster Generation Kit and Workflowfields in Record Details screen of Cluster Generation (Illumina SBS) 5.0 step
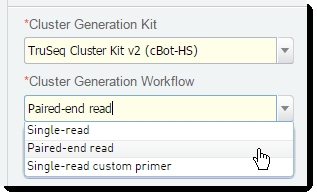
| • | In the Reagent Lot Tracking section, select the desired TruSeq Cluster Kit from the drop-down list. |
| • | In the Step Details area: |
| 3. | For Paired End Reads: In the Read 2 Cycles field, enter a value other than 0. |
| 4. | Enter values for the remaining required custom fields / UDFs. |
| 5. | Click the first button – Generate HiSeq Sample Sheet – to generate the sample sheet to be uploaded to the HiSeq/GAIIx instrument. |
| 6. | Once the Generate HiSeq Sample Sheet automation has completed, the HiSeq sample sheet and log file are generated and attached to the step. |
| • | Download the sample sheet file and upload to the HiSeq/GAIIx sequencing control software (HCS/SCS) to start a run. |
| 7. | Click the second button to generate the analysis sample sheet. In the screenshot below, the step has been configured to generate a bcl2fastq2 format analysis sample sheet (see Configuring Sample Sheet Generation). |
Figure 10: Configured sample sheet generation automations in Record Details screen of Cluster Generation (Illumina SBS) 5.0 step
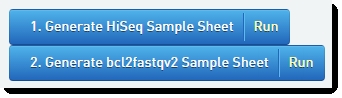
| 8. | Once the automation has completed: |
| • | bcl2fastq2: Download the sample sheet file; this will be used for analysis later in the workflow. |
| • | CASAVA: You do not need to download this file; it will be accessed during the Bcl Conversion and Demultiplexing (Illumina SBS) 5.0 step. |
| • | The analysis sample sheet and log file are generated and attached to the step. |
| 9. | (Optional) Upload the Lab Tracking Form to the appropriate placeholder. |
Figure 11: Record Details screen of Cluster Generation (Illumina SBS) 5.0 step (Files section)
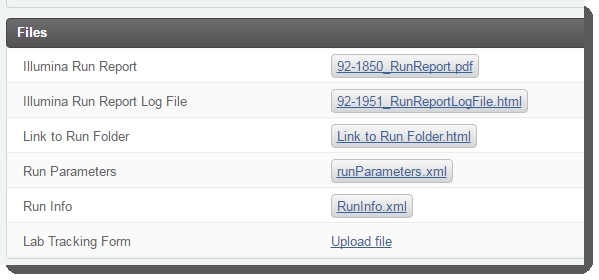
The Illumina Sequencing (Illumina SBS) 5.0 step records information for each lane of the flow cell and generates a report summarizing the results. In addition, run parameters, run info, the first base report (if generated), and a link to the run folder are automatically captured.
Table 3 describes the run information files, reports, placeholders, and links that the LIMS automatically generates or captures during a sequencing run.
Table 3: Run information generated or captured by Illumina Sequencing (Illumina SBS) 5.0 step
|
Item |
Description |
|
Run Info Run Parameters (HCS-controlled instruments only) RTA Configuration XML (SCS-controlled instruments only) |
These XML files are automatically captured by the LIMS from the instrument's run folder. They include the key run parameters, many of which are parsed out into key custom fields/UDFs. |
|
Link to Run Folder |
Automatically generated by the LIMS, this is a link to the network run folder where the data that was captured from the instrument during the run is stored. |
|
Illumina Run Report |
Automatically generated by the LIMS, this report provides key information about the run and the samples on the flow cell. Information includes the flow cell ID, run directory location, instrument name, as well as primary analysis metrics for the instrument – summarized per flow cell lane for the entire run, and individual reads in the case of paired-end runs. These metrics are compared against the instrument's per lane averages, calculated using metrics from the last 5 sequencing runs. Any values outside of 1 standard deviation are highlighted. |
|
First Base Report (HCS-controlled instruments only) |
Optionally generated during the instrument run by some Illumina instruments and captured by the LIMS, this report provides metrics to help determine the quality of the run. Note that manual file import is required for Sequencing Control Software (SCS)-controlled instruments. |
|
Lab Tracking Form placeholder |
This placeholder in the LIMS allows the user to manually attach a lab-specific tracking form to the step. |
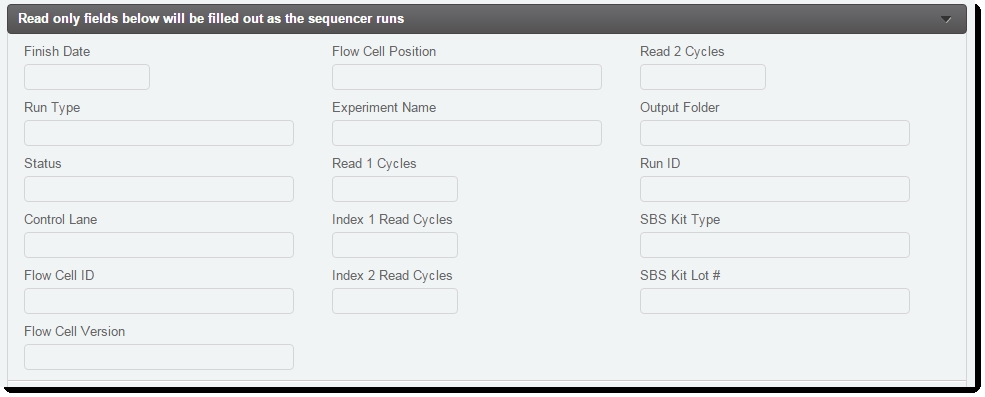
Listed below is the metadata that the LIMS automatically captures from the Illumina sequencing software as part of a sequencing run (see Figure 12). This information is gathered from various run result files and events.
| • | Date Run* – run start date |
| • | Instrument Run ID – sequencer instrument name |
| • | Control Lane – PhiX control lane (blank or 1-8) |
| • | Experiment Name – entered in software |
| • | Finish Date* – run completion date |
| • | Flow Cell ID |
| • | Flow Cell Position – A or B |
| • | Flow Cell Version |
| • | Output Folder – run folder root |
| • | Index 1 Read Cycles – intended Index cycles |
| • | Index 2 Read Cycles – intended Index cycles |
| • | Run ID – the unique run ID |
| • | Run Type – sequencing run type description |
| • | Status – current status of the sequencing run on the instrument |
| • | SBS Kit Lot # |
| • | SBS Kit Type |
Date Run is set to the date that the Begin Run event file is first picked up and associated with a step in BaseSpace Clarity LIMS, not the date of the Begin Run event itself (the date the run was performed on the instrument).
Finish Date is populated as follows:
| • | If the End Run event contains a date in the format YYYY-MM-DD, Finish Date will be set to the date in the event file |
| • | If the End Run event does not contain a date or the date is in the wrong format, Finish Date will be set to the date when the event file is processed |
Table 4 lists the Real Time Analysis (RTA) primary analysis metrics the LIMS automatically captures and records, per read, for samples in each flow cell lane. Some of these metrics are captured as soon as they become available during the run; others are captured upon run completion.
Table 4: RTA primary analysis metrics captured by Illumina Sequencing (Illumina SBS) 5.0 step
|
RTA primary analysis metrics |
Corresponding LIMS custom fields/UDFs |
|
Raw Yield (Gb) |
Yield PF (Gb) R1 Yield PF (Gb) R2 |
|
% Bases >Q30 |
% Bases >=Q30 R1 % Bases >=Q30 R2 |
|
Cluster Density (K/mm^2) |
Cluster Density (K/mm^2) R1 Cluster Density (K/mm^2) R2 |
|
Clusters Raw |
Clusters Raw R1 Clusters Raw R2 |
|
Clusters PF |
Clusters PF R1 Clusters PF R2 |
|
%PF |
%PF R1 %PF R2 |
|
Intensity Cycle 1 |
Intensity Cycle 1 R1 Intensity Cycle 1 R2 |
|
% Intensity Cycle 20 |
% Intensity Cycle 20 R1 % Intensity Cycle 20 R2 |
|
% Phasing |
% Phasing R1 % Phasing R2 |
|
% Prephasing |
% Prephasing R1 % Prephasing R2 |
|
% Aligned |
% Aligned R1 % Aligned R2 |
|
% Error Rate |
% Error Rate R1 % Error Rate R2 |
| 1. | The sequencing service runs in the background on the LIMS server. The service detects event files that the instrument software (RTA) is producing as the run progresses; this in turn lets the service know where to find the run data. |
| 2. | As the run data is written out and the events come in (Begin Run, Cycle Complete, End Run): |
| • | The data is matched to the step based on the flow cell ID, which was entered by the user as the Container Name on the Cluster Generation step. |
| • | Read-only custom field / UDF values on the Record Details screen are populated accordingly (see Figure 12 ). |
Figure 12: Record Details screen of Illumina Sequencing (Illumina SBS) 5.0 step showing custom fields/ UDFs used for result parsing
On the Record Details screen of the Illumina Sequencing (Illumina SBS) 5.0 step:
| 1. | The read-only custom field/UDF values are automatically populated as the sequencer runs. Figure 12 shows the Record Details screen fields that are used for result parsing. |
| 2. | Once the run has completed: |
| • | The read-only custom fields/UDFs shown in Figure 12 are populated. |
| • | The Illumina Run Report is generated and attached. |
| • | The Run Parameters and Run Info files are attached. |
| • | The Link to Run Folder is generated and attached. |
| • | QC flags are set on each lane. |
| 3. | [Optional] In theFiles section, upload the First Base Report and Lab Tracking Form. |
Figure 13: Record Details screen of Illumina Sequencing (Illumina SBS) 5.0 step (Files section)
Bcl Conversion and Demultiplexing
Invoking the Bcl conversion and demultiplexing automation triggers the following events:
| 1. | Initiate Bcl conversion with the correct parameters. |
| 2. | Parse Flowcell_demux_summary.xml . |
| 3. | Attach corresponding folder link file to each LIMS sample. |
| 4. | Parse sample stats. |
| 5. | Attach Demultiplex_Stats.htm and log file. |
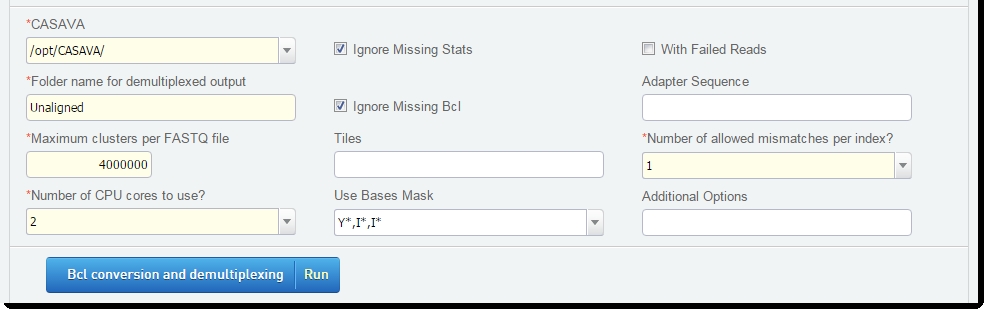
On the Record Details screen of the Bcl Conversion and Demultiplexing (Illumina SBS) 5.0 step:
| 1. | Review the Bcl parameters provided, and edit as required. |
| 2. | Click Bcl conversion and demultiplexing. |
Figure 14: Record Details screen of Bcl Conversion and Demultiplexing (Illumina SBS) 5.0 step
| 3. | Once the script has completed: |
| • | Demultiplex Stats and Script Log Details files are generated and attached to the step |
| • | HTML link files are attached to each demultiplexed sample |
| • | Parsed metrics are updated on the step |
| • | QC flags are set on the samples |
Setting the InstrumentType Parameter Value
If you are upgrading to the Illumina HiSeq/GAIIx Integration Package v5.5, you will need to set the value of the instrumentType parameter (-t) in the BCL Conversion and Demultiplexing step's EPP string.
This parameter value should be set to ‘hiseqga’, which is the instrument type prefix of the database properties used by the string.
The correctly configured parameter will look like this:
-t 'hiseqga’
In new installations of the Illumina HiSeq/GAIIx Integration Package v5.5, this parameter value is already set in the out-of-the-box configuration.
For details, see the following articles in the LIMS Integrations documentation:
| • | Normalization Buffer Volumes |
| • | Assign QC flags |
| • | CASAVA 1.8.2 Sample Sheet Generation |
| • | Bcl2fastq2 Sample Sheet Generation |
| • | BCL Conversion |
Components Installed with this Integration
The following sections describe the various components - files, database properties, reagent kits, and containers - that are installed by default as part of this integration. Information on installed protocols, steps, and automations is provided in the Protocols and automations section, above.
The Illumina HiSeq/GAII Integration is distributed as an RPM package that installs the components listed in Table 7. Note that this RPM package requires installation of the associated NGS Commons Package (see Release Notes).
Table 7: Illumina HiSeq/GAII Integration RPM package components
|
Component Installed |
Location and Description |
|
configure_extensions_illumina_hiseqgaii_workflow- v5.sh |
/opt/gls/clarity/config/ Script that imports the integration configuration. |
|
configure_extensions_hiseqgaii_sequencingservice- v5 .sh |
/opt/gls/clarity/config/ Script that imports the database properties for the integration. |
|
hcs-instrument-package- <version> ployment-bundle.zip |
/opt/gls/clarity/extensions/Illumina_HiSeq/v5/InstrumentIntegrations Instrument deployment package for HiSeq Control Software (HCS). Contains installation instructions and files to be deployed on the instrument. |
|
scs-instrument-package- <version> .sh -deployment-bundle.zip |
/opt/gls/clarity/extensions/Illumina_HiSeq/v5/InstrumentIntegrations Instrument deployment package for Sequencing Control Software (SCS). Contains installation instructions and files to be deployed on the instrument. |
|
log4j.properties, log4j.xml |
/opt/gls/clarity/extensions/Illumina_HiSeq/v5/SequencingService/conf File containing the settings for the sequencing jar’s logging. Note that as of HiSeq v. 5.4, the .properties file has been replaced by the XML file. |
|
libs directory, with multiple library jars |
/opt/gls/clarity/extensions/Illumina_HiSeq/v5/SequencingService Directory containing libraries used by the sequencing jar. Obsolete as of HiSeq v. 5.4, where these libraries are packaged with the jar file. |
|
hiseqgaii-extensions.jar |
/opt/gls/clarity/extensions/Illumina_HiSeq/<version>/EPP Jar file containing API-based LIMS extensions used throughout the workflow. |
|
hiseqgaii-sequencing.jar |
/opt/gls/clarity/extensions/Illumina_HiSeq/v5/SequencingService Sequencing service jar file that captures the run results. |
|
hiseqgaii_seqservice- v5.sh |
/opt/gls/clarity/extensions/Illumina_HiSeq/v5/SequencingService/bin Provides commands for interacting with the sequencing service. Accessible at /etc/init.d |
|
run_hiseqgaii_seqservice |
/opt/gls/clarity/extensions/Illumina_HiSeq/v5/SequencingService/bin Called by hiseqgaii_seqservice-v5, this script performs some basic system validation, then starts the sequencing service. |
Table 8 lists the database properties installed with the Illumina HiSeq/GAIIx Integration Package.
Note that several additional properties, each with the ‘99’ suffix appended to their name, are also installed. These are intended for use by the BaseSpace Clarity LIMS Support team in automated validation tests and are not listed in the table.
Constraints
Sequencing runs are matched using the flow cell ID and the sequencing step’s base name – Illumina Sequencing (Illumina SBS).
Do not change this base name – it is expected by the sequencing service that captures instrument run results. The base name is stored in the sequenceProcessBaseName database property (see Table 8). If this name is changed without the database property being updated, the 'flow cell ID <-> sequencing step base name' matching system will fail.
If required, you may modify the step name by editing or adding to the text after the base name portion as this is not used in the matching system. For example, you could change Illumina Sequencing (Illumina SBS) 5.0 to Illumina Sequencing (Illumina SBS) v5.
Table 8: Database properties installed with the Illumina HiSeq/GAIIx Integration Package
|
Property |
Description |
Default Value |
|
hiseqga.seqservice.ignoreUnmatchedContainerIds |
A flag indicating if event files that cannot be matched to flow cells in the LIMS should be archived after a certain period of time (true), or continually reprocessed (false). |
false To prevent the gls_events file directory from becoming cluttered, it is recommended that the value of this property is set to ‘true’. |
|
hiseqga.seqservice. ignoreUnmatchedContainerIdsWaitDays |
The number of days between when the event is created and the event file is archived. |
14 |
|
hiseqga.seqservice.synchronizationPeriod |
Invocation period in seconds. |
60 |
|
hiseqga.seqservice.sequenceProcessBaseName |
Sequencing process type base display name. Partial matching is used to look up the process type. |
Illumina Sequencing (Illumina SBS) |
|
hiseqga.seqservice.eventFileDirectory.1 |
A network location monitored for event files. |
/mnt/gls_events |
|
hiseqga.seqservice.netPathPrefixSearch.1 |
The network directory prefix contained in the event file. |
\\nas\network\run_data |
|
hiseqga.seqservice.netPathPrefixReplace.1 |
The mapped network directory mount name on the server used to access the run data directory. |
/mnt/run_data |
|
hiseqga.seqservice.eventFileDirectorySuffixes |
A list of eventFileDirectory path entries to monitor for event files. The value is one or more comma separated integers. |
99 |
|
hiseqga.seqservice.runReportViewsVersion |
The current version of the Run Report views in the database. The value 0 represents the state before the views are created. This property is automatically updated by the run report. |
0 |
|
hiseqga.seqservice.netPathPrefixSearchReplaceSuffixes |
A list of netPathPrefix search and replace entries for transforming Windows to Linux network paths. The value is one or more comma-separated integers. |
99 |
|
hiseqga.bcl.netPathPrefixSearch.1 |
(BCL conversion) The network directory prefix contained in the event file. |
\\nas\network\run_data |
|
hiseqga.bcl.netPathPrefixReplace.1 |
(BCL conversion) The mapped network directory mount name on the server used to access the run data directory. |
/mnt/run_data |
|
hiseqga.bcl.replacementSuffixes.bclserver |
(BCL conversion) A list of network path replacements associated with the BCL EPP Channel. The value is one or more comma-separated integers. The property format is hiseqga.bcl.replacementSuffixes.<EPP_CHANNEL_NAME> |
99 |
Support for multiple seqservice.netPathPrefixSearch property values
It is possible to configure support for multiple, identical seqservice.netPathPrefixSearch property values. For details, see Configuring Multiple Identical netPathPrefixSearch values.
The following reagent kits are included in the default configuration for the Illumina HiSeq/GAIIx Integration:
| • | AMPure XP Beads |
| • | TruSeq Cluster Kit |
| • | TruSeq Exome Enrichment - Box 1 |
| • | TruSeq Exome Enrichment - Box 2 |
The following control types are included in the default configuration for the Illumina HiSeq/GAIIx Integration:
| • | Endogenous Positive Control |
| • | Exogenous Positive Control |
| • | No Amplification Control |
| • | No Reverse Transcriptase Control |
| • | No Template Control |
| • | PhiX v3 |
The following container types are included in the default configuration for the Illumina HiSeq/GAIIx Integration:
| • | Illumina Flow Cell |
| • | Illumina Rapid Flow Cell |
To ensure that your Illumina instrument warranty remains valid, the instrument integration must be performed and maintained by the BaseSpace Clarity LIMS Support team. To perform this integration, the Support team will require direct access to the instrument via WebEx or Remote Desktop while the instrument is idle.
Below is an overview of the steps performed by the LIMS support team when configuring the instrument for use with the Illumina HiSeq/GAIIx Integration v5.5 to BaseSpace Clarity LIMS.
Configuring the Illumina Instrument
| 1. | Create directory on the local computer to hold the batch files. These batch files write event files to the network attached storage (NAS) share(s). |
| 2. | Create directory on the NAS to hold the event files. |
| 3. | Modify Illumina software configuration files to call the batch files that create the event files. |
| 4. | Update sequencing service default database properties to match the specifics of the installation. |
We recommend configuring sequencer instrument names in the LIMS to track the specific lab instrument on which a run was completed.
This information should be c onfigured on the Illumina Sequencing(Illumina SBS)5.0 step, and should be set to the same name as is configured in the Illumina sequencing software – i.e., this will be value of the Instrument Run ID field described in the Metadata section above.
When the integration reads the Instrument Run ID field in the run results, it will update the instrument on the step in the LIMS, eliminating the need for the user to manually enter this information.
For instructions on configuring instrument names, refer to the following articles:
LIMS v4.1 and v4.2 Documentation:
| • | Adding Instruments and Instrument Types (System Configuration section) |
LIMS v5 Documentation:
| • | Adding and Configuring Instruments (Consumables section) |
Configuring Sample Sheet Generation
Sample sheet generation occurs on the step prior to the sequencing run – Cluster Generation (Illumina SBS) 5.0 – which is the step where samples are placed on the container that will be loaded in the instrument.
The out-of-the-box configuration provides three automations (listed below in Table 3). These generate the following:
| • | One sample sheet for use with the HiSeq/GAII(x) instrument sequencing control software (CASAVA v1.8.2 format). |
| • | Two options for an analysis sample sheet for use with CASAVA v1.8.2 or bcl2fastq v2.15 downstream analysis – at least one of which should be disabled when the system is initially configured (instructions provided below ). If you do not require a sample sheet for use in downstream analysis, you may prefer to disable both options. |
By default, one instrument and one analysis sample sheet are created for the flow cell loaded during the Cluster Generation (Illumina SBS) 5.0 step.
If required, you may configure the step to generate analysis sample sheets for multiple flow cells. For details, refer to the sample sheet generation article for your analysis software:
| • | CASAVA 1.8.2 Sample Sheet Generation |
| • | Bcl2fastq2 Sample Sheet Generation (NGS v5.16 and earlier) |
The sample sheet generation automations are configured on the Record Details screen of the Cluster Generation (Illumina SBS) 5.0step.
Figure 15: Record Details screen of Cluster Generation step showing out-of-the-box sample sheet generation automations prior to configuration
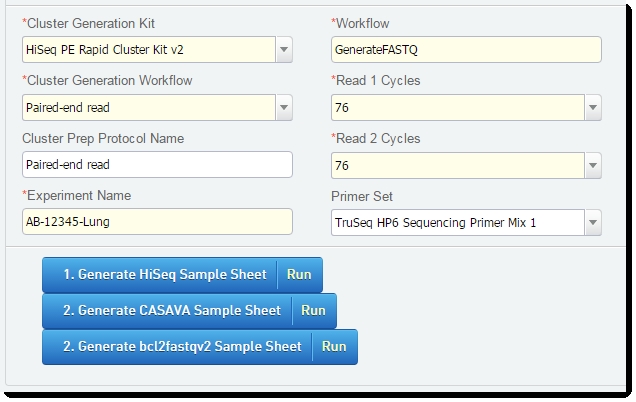
Table 5 shows the format of the sample sheets generated by each of these automations.
Table 5: Sample sheet generation automations and file formats
|
Automation |
Generated Sample Sheet Format |
|
1. Generate HiSeq Sample Sheet |
CASAVA v1.8.2 |
|
2. Generate CASAVA Sample Sheet |
CASAVA v1.8.2 |
|
2. Generate bcl2fastqv2 Sample Sheet |
bcl2fastq v2.15 |
Upgrading Cluster Generation (Illumina SBS) 5.0 Master Step Custom Fields/Step UDFs
If you are upgrading to the Illumina HiSeq/GAIIx Integration Package v5.5, note the following:
| • | To enable generation of the bcl2fastq2 sample sheet, additional fields / UDFs have been added to the Cluster Generation (Illumina SBS) 5.0 step. These are listed in Table 6. |
Table 6: Cluster Generation (Illumina SBS) 5.0 - additional master step fields/step UDFs
|
Name |
Type |
Field Constraints |
Preset Values |
|
Adapter |
Single-line Text |
Required |
|
|
Adapter Read 2 |
Single-line Text |
||
|
Read 1 Cycles |
Numeric |
Required, minimum value 0 |
151, 76, and 51 |
|
Read 2 Cycles |
Numeric |
Required, minimum value 0 |
151 and 76 |
|
Experiment Name |
Single-line Text |
Required |
|
|
Workflow |
Single-line Text |
Required Use first Preset Value as the default value option selected |
GenerateFASTQ |
| • | The TruSeq Cluster Generation Kit master step field/step UDF has been renamed Cluster Generation Kit and its preset values have been updated as follows: |
| – | TruSeq Cluster Kit v2 (cBot-HS) |
| – | TruSeq Cluster Kit v3 (cBot-HS) |
| – | HiSeq PE Cluster Kit v4 |
| – | HiSeq SR Rapid Cluster Kit v2 |
| – | HiSeq PE Rapid Cluster Kit v2 |
| • | The master step fields / step UDFs Cluster Generation Kit and Cluster Generation Workflow are now required. |
Disable bcl2fastqv2 and Use the CASAVA Sample Sheet for Analysis
LIMS 4.x
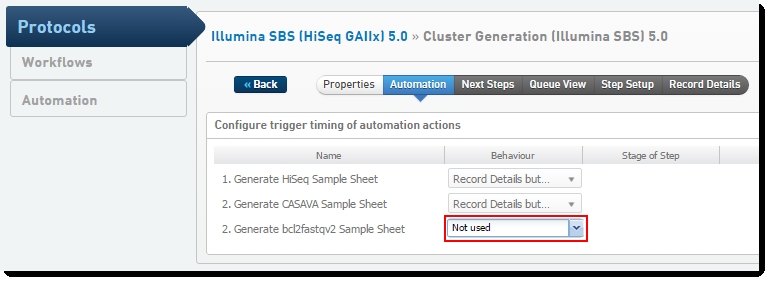
| 1. | In the LIMS Web Interface, on the Configuration screen, click Protocols. |
| 2. | Edit the Illumina SBS (HiSeq GAIIx) 5.0 protocol. |
| 3. | Edit the Cluster Generation (Illumina SBS) 5.0 step. |
| 4. | On the Automation tab, set the value for 2. Generate bcl2fastqv2 Sample Sheet to "Not used." |
Figure 16: Disabling generation of the bcl2fastqv2 sample sheet
| 5. | Save the step configuration. |
LIMS 5.x
| 1. | In the LIMS, click the Configuration menu item and then click the Lab Work tab. |
| 2. | On the Lab Work configuration screen, in the Workflows, Protocols, Steps, and Master Steps navigation panel, select (click) the Illumina SBS (HiSeq GAIIx) 5.0 protocol. The Steps and Master Steps lists update, highlighting the steps included in the protocol, and the master steps on which those steps are based. |
| 3. | Select the Cluster Generation (Illumina SBS) 5.0step. |
Figure 17: Selecting the Cluster Generation (Illumina SBS) 5.0 step
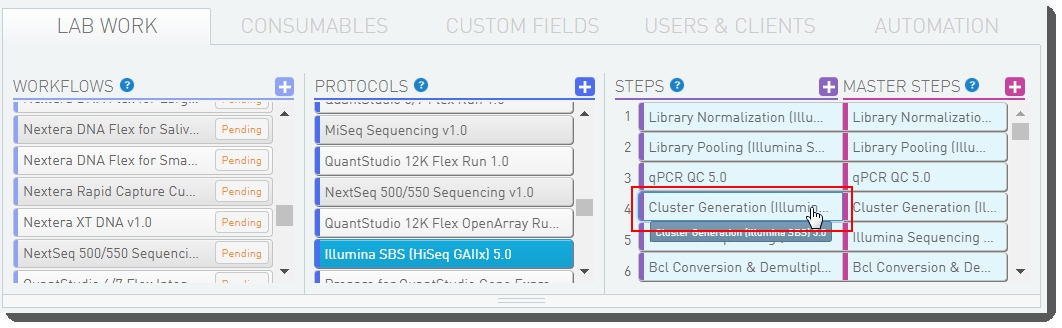
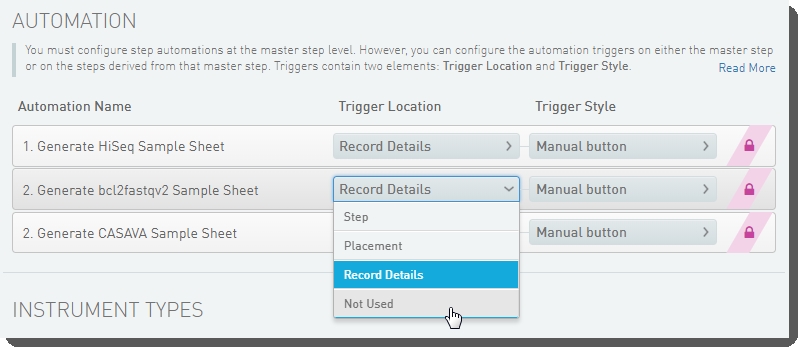
| 4. | Below the navigation panel, the step and master step configuration forms display. Scroll down to the Automation configuration section. For the 2. Generate bcl2fastqv2 Sample Sheet automation, set the Trigger Location to Not Used. |
Figure 18: Disabling generation of the bcl2fastqv2 sample sheet
| 5. | Save the step configuration. |
After completing the configuration steps above, when the step is run, only two buttons will be visible in Record Details view.
Note that both generate the same CASAVA format sample sheet.
Figure 19: Automations configured for use with HiSeq/GAII and CASAVA downstream analysis

Disable CASAVA and Use the bcl2fastq2 Sample Sheet for Analysis
LIMS 4.x
| 1. | In the LIMS Web Interface, on the Configuration screen, click Protocols . |
| 2. | Edit the Illumina SBS (HiSeq GAIIx) 5.0 protocol. |
| 3. | Remove the BCL Conversion and Demultiplexing (Illumina SBS) 5.0 step. |
Figure 20: Removing BCL Conversion and Demultiplexing from Illumina SBS (HiSeq GAIIx) protocol
| 4. | Save the protocol configuration. |
| 5. | Edit the Cluster Generation (Illumina SBS) 5.0 step. |
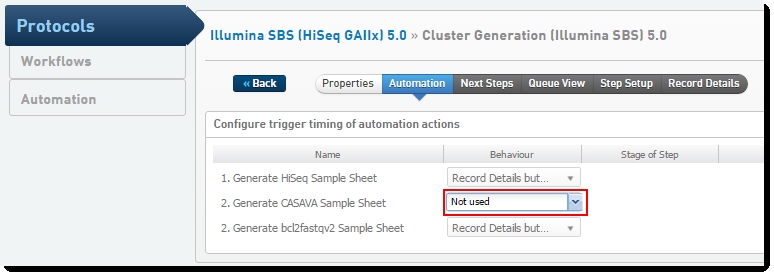
| 6. | On the Automation tab, set the value for 2. Generate CASAVA Sample Sheet to "Not used." |
Figure 21: Disabling generation of the CASAVA sample sheet
| 7. | Save the step configuration. |
LIMS 5.x
| 1. | In the LIMS, click the Configuration menu item and then click the Lab Work tab. |
| 2. | On the Lab Work configuration screen, in the Workflows, Protocols, Steps, and Master Steps navigation panel, select (click) the Illumina SBS (HiSeq GAIIx) 5.0 protocol. The Steps and Master Steps lists update, highlighting the steps included in the protocol, and the master steps on which those steps are based. |
| 3. | Select the BCL Conversion and Demultiplexing (Illumina SBS) 5.0 step and click Delete. |
Figure 22: Removing BCL Conversion and Demultiplexing from Illumina SBS (HiSeq GAIIx) 5.0 protocol
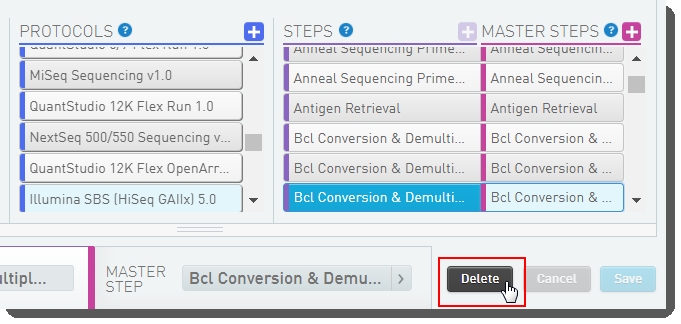
| 4. | Save the protocol configuration. |
| 5. | In the Steps list, select the Cluster Generation (Illumina SBS) 5.0step. |
Figure 23: Selecting the Cluster Generation (Illumina SBS) 5.0 Step

| 6. | Below the navigation panel, the step and master step configuration forms display. Scroll down to the Automation configuration section. For the 2. Generate CASAVA Sample Sheet automation, set the Trigger Location to Not Used. |
Figure 24: Disabling Generation of the CASAVA Sample Sheet
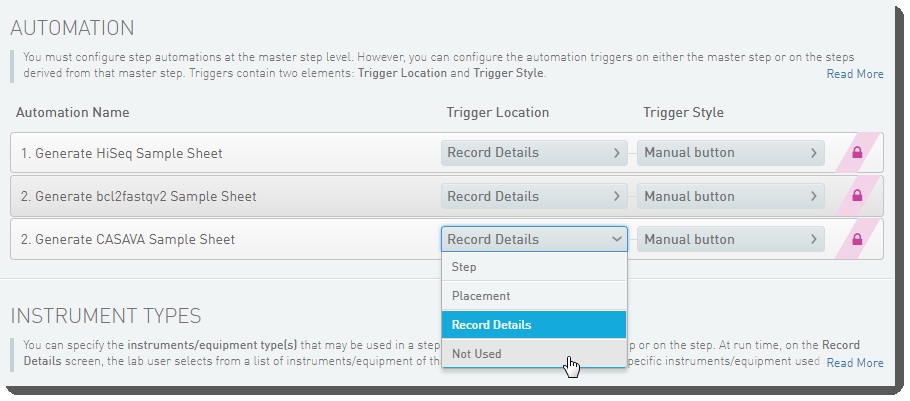
| 7. | Save the step configuration. |
After completing the configuration steps above, when the step is run, only two buttons will be visible in Record Details view.
Figure 25: Automations configured for use with HiSeq/GAIIx and bcl2fastq2 downstream analysis

See the CASAVA 1.8.2 Sample Sheet Generation and Bcl2fastq2 Sample Sheet Generation (NGS v5.16 and earlier) articles for detailed information on:
| • | Sample sheet generation script parameters and usage |
| • | Sample sheet data and configuration options |
| • | Enabling sample sheet generation for multiple flow cells |
| • | Enabling unique FASTQ file names per sequencing run |
| • | Sample sheet rules and constraints |
Support for container types
All one-dimensional container types with both numeric rows and numeric columns are supported.
Rules and Constraints
This integration operates with the following constraints:
| • | The flow cell ID must be unique. There should not be multiple flow cell containers in the system with the same name. |
| • | The flow cell ID must be scanned as the flow cell Container Name on the Cluster Generation (Illumina SBS) 5.0 step. |
| • | The sample sheet provided in the BaseCalls directory must be named SampleSheet.csv. |
| • | For sample sheet generation constraints, refer to the appropriate article for your analysis software: |
| – | CASAVA 1.8.2 Sample Sheet Generation |
| – | Bcl2fastq2 Sample Sheet Generation (NGS v5.16 and earlier) |
Vendor Resources
| • | Illumina HiSeq 2500 System information |
| • | Guided sample sheet creation and setup |
| • | Illumina Experiment Manager User Guide |
| • | Illumina Experiment Manager Support |
