NovaSeq 6000 Integration v3.2.1 User and Configuration Guide
The Illumina NovaSeq 6000 Integration Package v3.2.1 supports the integration of BaseSpace Clarity LIMS to Illumina NovaSeq 6000 sequencers.
Note the following key changes for this version of the integration:
| • | A move from a file-based to an API-based integration using the Sequencer API. |
| • | NovaSeq-specific configuration is now installed as part of the Preconfigured Workflows Package (PCWP) v1.2. This configuration includes new template path locations and sample sheet generation with Template File Generator (TFG). |
Configuration changes in v3.2.1 include the following:
| • | Support for Sequencer API |
| • | Support for SPrime (SP) flow cell |
| • | Support for Long-Read sequencing (SP flow cell only) |
| • | Support for asymmetric read lengths |
This User and Configuration Guide describes the integration and includes information about the preconfigured workflows, protocols, steps, and automations. The guide also lists installed components, and provides configuration requirements, rules, and constraints.
For instructions on validating and troubleshooting the Illumina NovaSeq 6000 Integration, see the NovaSeq 6000 Integration v3.2.1 Validation and Troubleshooting Guide.
The configuration provided in this integration has been established to support NovaSeq 6000 lab processes. Any configuration changes to protocols or workflows - including renaming protocols, steps, and fields - could break process.
The integration includes the following:
| • | Preconfigured NovaSeq 6000 v3.0 workflow that maps to lab protocols and instrument runs. |
| • | Preconfigured protocols: |
| – | Run Format (NovaSeq 6000 v3.0) |
| – | NovaSeq Standard (NovaSeq 6000 v3.0) (supports the loading of pooled libraries into a library tube) |
| – | NovaSeq Xp (NovaSeq 6000 v3.0) (supports individual lane loading on the NovaSeq) |
| – | AUTOMATED - NovaSeq Run (NovaSeq 6000 v3.0) |
| • | Automated generation of a sample sheet for use with bcl2fastq2 v2.20 analysis software. This file is automatically uploaded to the sequencing instrument via the Sequencer API and is used to set up the run. |
| • | Automatic validation of run setup information. This information is used to create the run recipe and initiate the run. |
| • | Automated tracking of the NovaSeq sequencing run and parsing of run statistics into the LIMS, including: |
| – | Progress and metrics of sequencing run |
| – | Per-instrument sequencing runs (tracked as part of the Run ID field value) |
| – | Sequencing run parameters |
| – | Sequencing run data directory location |
| – | Real Time Analysis v3 (RTA3) version and other run specific information |
| • | [Optional] Preconfigured Library Prep Validation v1.0 workflow used for validation purposes only. The workflow contains a single-step protocol that models the library prep required to produce normalized libraries that are ready for the NovaSeq 6000 v3.0 workflow. See the NovaSeq 6000 Integration v3.2.1 Validation and Troubleshooting Guide. |
Information on these features is provided in the following sections.
It is assumed that samples enter the NovaSeq 6000 v3.0 workflow as normalized libraries. That is, before they are assigned to the workflow:
| • | Samples have been accessioned into the LIMS. |
| • | Samples have been run through QC and library prep. |
| • | Samples have been normalized, and the value is captured in a field called Normalized Molarity (nM). |
For details on sample accessioning, see the following articles in the Samples section of the LIMS documentation for your version: LIMS Documentation v5.0 and v5.1 or LIMS Documentation v4.1 and v4.2:
| • | Sample Accessioning |
| • | Using Sample Lists to Upload and Modify Samples |
Samples can be assigned to the NovaSeq 6000 v3.0 workflow automatically (using a routing script), or manually from the Projects and Samples dashboard (see the Assigning Samples to Workflows article in the LIMS Documentation).
Workflows, Protocols, and Steps
The Illumina NovaSeq 6000 Integration Package v3.2.1 includes two workflows:
| • | NovaSeq 6000 v3.0 |
| • | Library Prep Validation v1.0 (optional, but recommended for validation purposes) |
The following table lists and describes the protocols and steps included in these workflows.
|
Library Prep Validation v1.0 workflow |
|||||||||||||||||
|
Protocol name |
Protocol purpose |
Steps included in protocol |
|||||||||||||||
|
Library Prep Validation v1.0 |
Included for validation purposes only, this protocol models the library prep steps required to advance samples to the Run Format (NovaSeq 6000 v3.0) protocol. The protocol contains a single step - Library Prep Validation v1.0. At the end of this step, a routing script sends the samples to the first step of the NovaSeq 6000 v3.0 workflow - Define Run Format (NovaSeq 6000 v3.0). |
|
|||||||||||||||
|
NovaSeq 6000 v3.0 workflow |
|||||||||||||||||
|
Protocol name |
Protocol purpose |
Steps included in protocol |
|||||||||||||||
|
Run Format (NovaSeq 6000 v3.0) |
Allows for the assignment of per sample values for Loading Workflow Type, Normalized Molarity, Flowcell Type, and Final Loading Concentration (pM):
Compares each sample's Normalized Molarity value with the Minimum Molarity value. Routing script sends samples to the NovaSeq Standard or NovaSeq Xp protocol, according to the selected Loading Workflow Type. Samples with Normalized Molarity less than Minimum Molarity are removed from the workflow. |
|
|||||||||||||||
|
NovaSeq Standard (NovaSeq 6000 v3.0) |
Samples are pooled and added to the library tube in preparation for the NovaSeq run. Run setup information is validated and sample sheet is generated. Routing script sends library tube to the AUTOMATED - NovaSeq Run (NovaSeq 6000 v30) protocol. |
|
|||||||||||||||
|
NovaSeq Xp (NovaSeq 6000 v3.0) |
Samples are pooled and added to lanes on the NovaSeq flow cell. The flow cell type is determined by the option selected in the Define Run Format (NovaSeq 6000 v3.0) step. Run setup information is validated and sample sheet is generated. Routing script sends flow cell to the AUTOMATED - NovaSeq Run (NovaSeq 6000 v30) protocol. |
|
|||||||||||||||
|
AUTOMATED - NovaSeq Run (NovaSeq 6000 v3.0) |
All samples complete the workflow by going through this protocol. This protocol contains one fully-automated step. The user must not add samples to the Ice Bucket or start the step. The integration will do this automatically. |
|
|||||||||||||||
The following image show the protocols and their steps displayed in the LIMS Lab View.
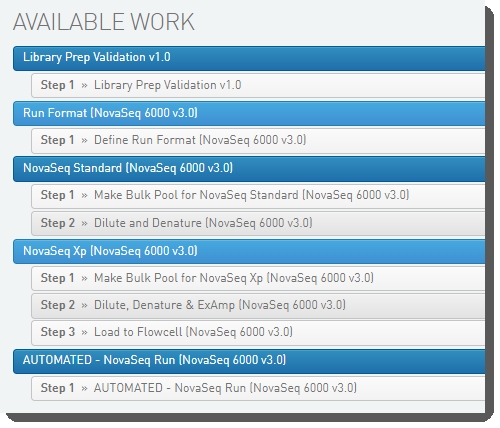
The Library Prep Validation v1.0 workflow allows for validation of the system after installation is complete. For details, see the NovaSeq 6000 Integration v3.2.1 Validation and Troubleshooting Guide.
The following table lists the step automations included in the NovaSeq 6000 v3.0 workflow. Automation details are provided in the sections following the table.
For details on the validation workflow, see the NovaSeq 6000 Integration v3.2.1 Validation and Troubleshooting Guide.
|
Protocol |
Step |
Automation |
||||||||||||
|
1. Run Format (NovaSeq 6000 v3.0) |
1. Define Run Format (NovaSeq 6000 v3.0) |
|
||||||||||||
|
2. NovaSeq Standard (NovaSeq 6000 v3.0) |
1. Make Bulk Pool for NovaSeq Standard (NovaSeq 6000 v3.0) |
|
||||||||||||
|
2. Dilute and Denature (NovaSeq 6000 v3.0) |
|
|||||||||||||
|
3. NovaSeq Xp (NovaSeq 6000 v3.0) |
1. Make Bulk Pool for NovaSeq Xp (NovaSeq 6000 v3.0) |
|
||||||||||||
|
2. Dilute, Denature and ExAmp (NovaSeq 6000 v3.0) |
|
|||||||||||||
|
3. Load to Flowcell (NovaSeq 6000 v3.0) |
|
|||||||||||||
|
4. AUTOMATED - NovaSeq Run (NovaSeq 6000 v3.0) |
1. AUTOMATED - NovaSeq Run (NovaSeq 6000 v3.0) |
|
The sections below describe the configuration installed with the Illumina NovaSeq 6000 Integration.
As of LIMS v5, the term user-defined field (UDF) has been replaced with custom field in the user interface (the API resource is still called udf). There are two types of custom fields:
| • | Master step fields - configured on master steps. These fields only apply to the master step on which they are configured, and the steps derived from those master steps. |
| • | Global fields - configured on entities such as submitted sample, derived sample, measurement, etc. These fields apply to the whole LIMS system. |
Protocol 1: Run Format (NovaSeq 6000 v3.0
This protocol sets the Loading Workflow Type and allows the user to choose the appropriate Flowcell Type and Final Loading Concentration (pM). At the end of the protocol, a routing script sends the normalized libraries to either the NovaSeq Standard (NovaSeq 6000 v3.0) or the NovaSeq Xp (NovaSeq 6000 v3.0) protocol.
This protocol contains one step: Define Run Format (NovaSeq 6000 v3.0), described below.
Step 1. Define Run Format (NovaSeq 6000 v3.0)
Step input: NTP (normalized libraries)
Step output: None
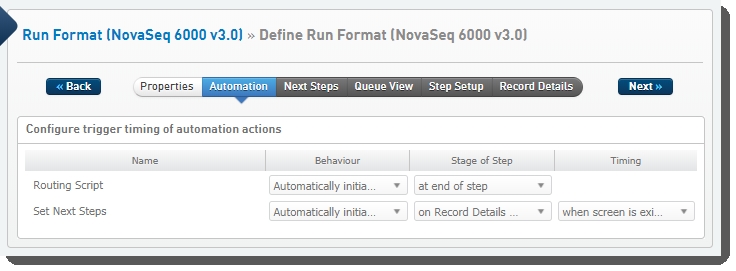
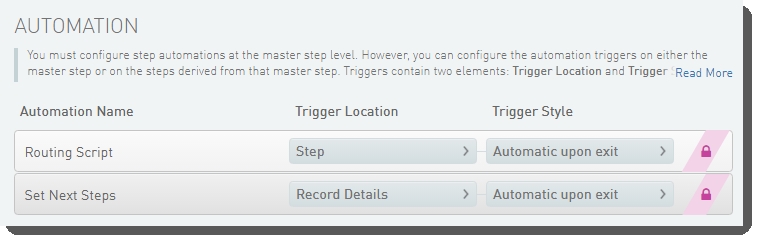
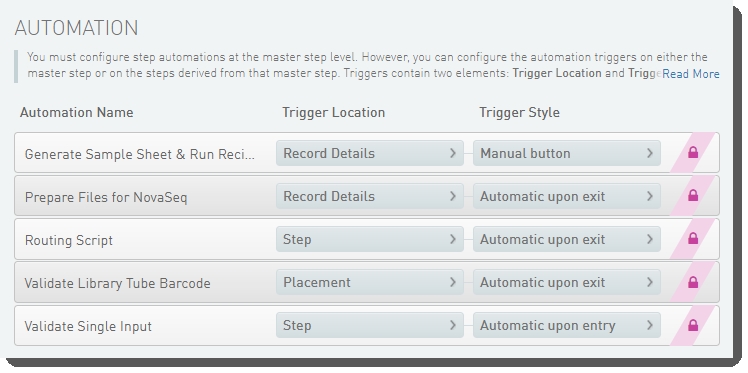
The following automations are configured on this step:
| • | Routing Script |
| • | Set Next Steps |
Define Run Format (NovaSeq 6000 v3.0) automation configuration - LIMS v4.2.x:
Define Run Format (NovaSeq 6000 v3.0) automation configuration - LIMS v5.x:
These automations are described below, in the order in which they are triggered at run time.
Set Next Steps
Automatically triggered on exit of the Record Details screen, this automation does the following:
| • | Sets the next step for samples to REMOVE: |
nextStep = ::REMOVE::
| • | Calculates the Minimum Molarity using the following formula: |
input.::Minimum Molarity (nM):: = (5 * input.::Final Loading Concentration (pM)::)/1000
| • | Checks Normalized Molarity value. For samples with no Normalized Molarity value (i.e., empty value, not including 0), generates an error message informing the user that the field cannot be empty: |
if (!input.hasValue(::Normalized Molarity (nM)::)) { \
fail(::The Normalized Molarity cannot be empty.::) ;
}
| • | Compares each sample's Normalized Molarity value with the Minimum Molarity value. If Normalized Molarity value is lower than the Minimum Molarity value, sets the sample's Loading Workflow Type to [Remove from workflow] and records a message in the Warning field for the sample: |
else if (input.::Normalized Molarity (nM):: < input.::Minimum Molarity (nM)::) { \
input.::Warning:: = ::The Normalized Molarity is too low.:: ; \
input.::Loading Workflow Type:: = ::[Remove from workflow]:: ; } \
else { input.::Warning:: = ::n/a:: }
At this point the user has two options:
| – | Correct the Normalized Molarity value on the Record Details screen. User will also need to edit the Loading Workflow Type field and set it to NovaSeq Standard or NovaSeq Xp, as applicable. |
| – | Complete the protocol without correcting the Normalized Molarity value. In this case, those samples will be removed from the workflow. |
Routing Script
Automatically triggered on exit of the step, this automation invokes the changeWorkflow script, which routes step inputs appropriately.
| • | Samples with Loading Workflow Type field value = NovaSeq Standard are routed to the NovaSeq 6000 v3.0 workflow and queued for the Make Bulk Pool for NovaSeq Standard (NovaSeq 6000 v3.0) step. |
| • | Samples with Loading Workflow Type field value = NovaSeq Xp are routed to the NovaSeq 6000 v3.0 workflow and queued for the Make Bulk Pool for NovaSeq Xp (NovaSeq 6000 v3.0) step. |
Default automation command line:
bash -c "/opt/gls/clarity/bin/java -jar /opt/gls/clarity/extensions/ngs-common/v5/EPP/ngs-extensions.jar -u {username} -p {password} -i {stepURI:v2} -l {compoundOutputFileLuid0} script:changeWorkflow \\
--FIELD_NAME 'Loading Workflow Type' \
--FIELD_VALUE 'NovaSeq Standard' \
--WORKFLOW 'NovaSeq 6000 v3.0' \
--STEP 'Make Bulk Pool for NovaSeq Standard (NovaSeq 6000 v3.0)' \
--INPUTS_OR_OUTPUTS 'INPUTS' \\
--FIELD_NAME 'Loading Workflow Type' \
--FIELD_VALUE 'NovaSeq Xp' \--WORKFLOW 'NovaSeq 6000 v3.0' \
--STEP 'Make Bulk Pool for NovaSeq Xp (NovaSeq 6000 v3.0)' \
--INPUTS_OR_OUTPUTS 'INPUTS'"
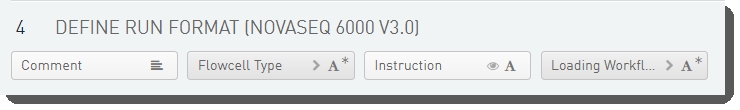
Step UDFs/Custom Fields
There are four fields defined on the Define Run Format (NovaSeq 6000 v3.0) step.
These are shown below in the BaseSpace Clarity LIMS v5.x Configuration area (Custom Fields > Master Step Fields tab).
See the following table for LIMS v4.2.x and LIMS v5.x field configuration details:
|
Field Name |
Field Type |
Field Constraints |
Preset Values/Additional Options and Dropdown Items |
|||||||||||||||||||||
|
Comment |
Multi-line Text |
|
None |
|||||||||||||||||||||
|
Flowcell Type |
Single-line Text (v4.2.x) Text Dropdown (v5.x) |
|
|
|||||||||||||||||||||
|
Instruction |
Single-line Text (v4.2.x) Text (v5.x) |
|
|
|||||||||||||||||||||
|
Loading Workflow Type |
Single-line Text (v4.2.x) Text Dropdown (v5.x) |
|
|
Global UDFs/Custom Fields
The following table lists the global UDFs/custom fields that are configured to display on the Define Run Format (NovaSeq 6000 v3.0) step.
|
Field Name |
Field Type |
Field Constraints/Options |
Preset Values/Additional Options and Dropdown Items |
|||||||||||||||||||||
|
Configured on Analyte in LIMS v4.2.x and Derived Samplein LIMS v5.x: |
||||||||||||||||||||||||
|
Adjusted Per Sample Volume (ul) |
Numeric |
|
|
|||||||||||||||||||||
|
Final Loading Concentration (pM) |
Numeric (v4.2.x) Numeric Dropdown (v5.x) |
|
|
|||||||||||||||||||||
|
Flowcell Type |
Single-line Text (v4.2.x) Text Dropdown (v5.x) |
|
|
|||||||||||||||||||||
|
Loading Workflow Type |
Single-line Text (v4.2.x) Text Dropdown (v5.x) |
|
|
|||||||||||||||||||||
|
Minimum Molarity (nM) |
Numeric |
|
|
|||||||||||||||||||||
|
Normalized Molarity (nM) |
Numeric |
|
|
|||||||||||||||||||||
|
Per Sample Volume (ul) |
Numeric |
|
|
|||||||||||||||||||||
|
Warning |
Single-line Text (v4.2.x) Text Dropdown (v5.x) |
|
|
|||||||||||||||||||||
User Interaction
| 1. | On the Record Details screen, the Sample Details table contains the following fields that must be completed (values can vary across samples): |
| • | Loading Workflow Type: Users selects NovaSeq Standard or NovaSeq Xp from the drop-down list. |
| • | Flowcell Type: User selects SP, S1, S2, or S4 from the drop-down list. |
| • | Final Loading Concentration (pM): User selects from the two preset options - 225 (for PCR-free workflows) or400 (for Nano workflows), or may enter a different value. |
| • | Normalized Molarity (nM):These values are copied over from the previous step. If the user did not populate this column during library prep, they can enter the values here. |
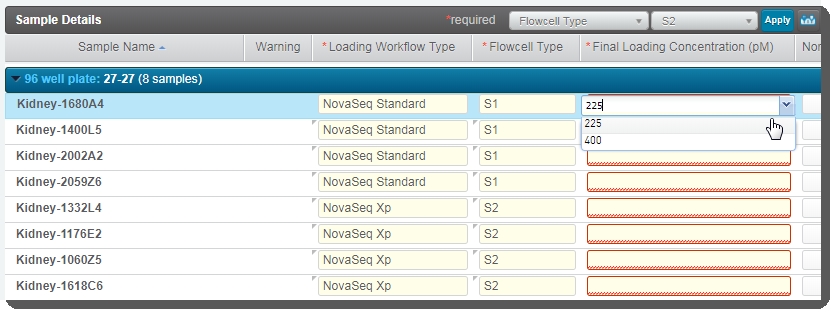
| • | User clicks Next Steps. This triggers the Next Steps automation, which does the following: |
| – | Sets the value of the next step (for all samples) to Remove from workflow. The Routing Script automation expects this value, and requires it in order to successfully advance samples to the next step. |
| – | Calculates the Minimum Molarity. |
| – | Checks Normalized Molarity value. For samples with no Normalized Molarity value (i.e., empty value, not including 0), generates an error message informing the user that the field cannot be empty. |
| – | Compares each sample's Normalized Molarity value with the Minimum Molarity value. |
| 2. | On the Assign Next Steps screen: |
| • | In the Sample Details table, the Next Step for all samples is pre-populated with Remove from workflow - regardless of the Loading Workflow Type. |
The user must not change this value. If Next Step is not set to Remove from workflow, the routing script will not be able to route samples correctly.
For samples whose Normalized Molarity value was found to be lower than the Minimum Molarity value, the Loading Workflow Type is set to Remove from workflow and a message is recorded in the Warningfield for the sample:
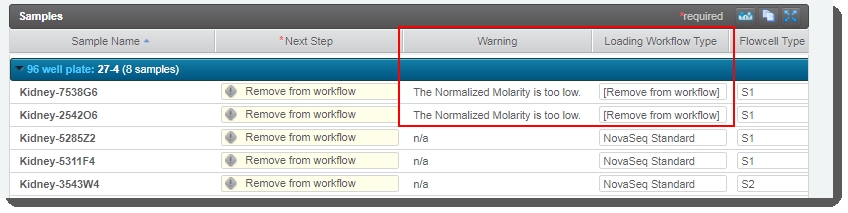
At this point the user has two options:
| – | Return to the Record Details screen and adjust the Normalized Molarity value so that it equals or exceeds the Minimum Molarity value. User will also need to set the Loading Workflow Type to NovaSeq Standard. |
| – | Complete the protocol without correcting the Normalized Molarity value. In this case, the samples in question will be removed from the LIMS workflow. |
| 3. | User clicks Finish Step. The Routing Script automation is triggered: |
| • | Samples whose Loading Workflow Type is set to Remove from workflow (i.e., whereNormalized Molarity value is lower than the Minimum Molarity) are removed from the LIMS workflow. |
| • | Samples whose Loading Workflow Type is set to NovaSeq Standard are routed to the Make Bulk Pool for NovaSeq Standard (NovaSeq 6000 v3.0) step. This is the first of two steps in the NovaSeq Standard (NovaSeq 6000 v3.0) protocol. |
| • | Samples whose Loading Workflow Type is set to NovaSeq Xp are routed to the Make Bulk Pool for NovaSeq Xp (NovaSeq 6000 v3.0) step. This is the first of three steps in the NovaSeq Xp (NovaSeq 6000 v3.0) protocol. |
Protocol 2: NovaSeq Standard (NovaSeq 6000 v3.0)
Samples are routed to this protocol if their Loading Workflow Type value is set to NovaSeq Standard. Samples are pooled and added to a library tube in preparation for the NovaSeq run.
At the end of this protocol, a routing script sends the library tube to the AUTOMATED - NovaSeq Run (NovaSeq 6000 v3.0) protocol.
This protocol contains two steps, described below:
| • | Step 1: Make Bulk Pool for NovaSeq Standard (NovaSeq 6000 v3.0) |
| • | Step 2: Dilute and Denature (NovaSeq 6000 v3.0) |
Step 1: Make Bulk Pool for NovaSeq Standard (NovaSeq 6000 v3.0)
In this step, user manually places libraries into a single pool. Resuspension buffer and reagents are added.
User must only create only one pool per step.
Step input: NTP (normalized libraries)
Step output: Bulk pool
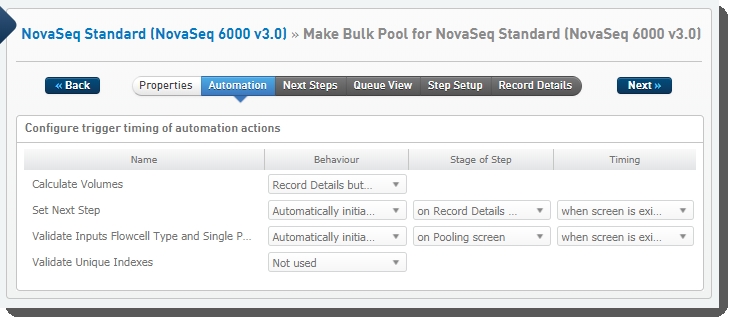
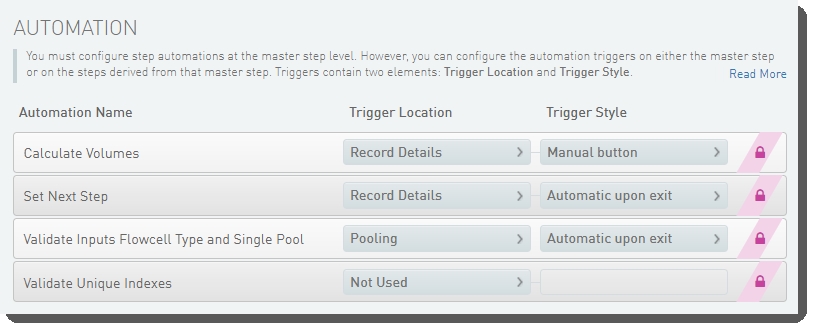
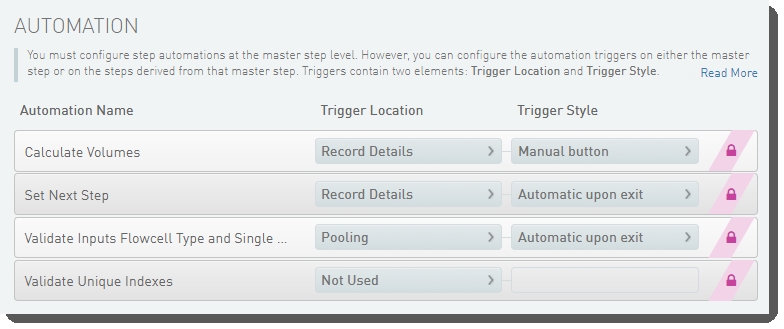
The following automations are configured on the step:
| • | Calculate Volumes |
| • | Set Next Step |
| • | Validate Inputs Flowcell Type and Single Pool |
| • | Validate Unique Indexes(Not used. This functionality is handled by the core LIMS configuration for pooling.) |
Make Bulk Pool for NovaSeq Standard (NovaSeq 6000 v3.0) automation configuration - LIMS v4.2.x:
Make Bulk Pool for NovaSeq Standard (NovaSeq 6000 v3.0) automation configuration - LIMS v5.x:
These automations are described below, in the order in which they are triggered at run time.
Validate Inputs Flowcell Type and Single Pool
Automatically triggered on exit of the Pooling screen, this automation:
| • | Checks that all samples in the pool have the same Flowcell Type assigned to them. |
| • | Checks that only one pool has been created. Changes the value of error and logging messages to reference the type of pool passed as 'bulk' pool. |
/opt/gls/clarity/bin/java -jar /opt/gls/clarity/extensions/ngs-common/v5/EPP/ngs-extensions.jar \script:validate_flowcell_for_input_pools \
-i {stepURI:v2} \
-u {username} \
-p {password} \
-l {compoundOutputFileLuid1} \
-validateSingleOutput true \
-poolType bulk"
Calculate Volumes
Automatically triggered when the user clicks the Calculate Volumes button on the Record Details screen, this automation:
| • | Calculates the number of samples in the pool: |
-exp 'step.::Number of Samples in Pool:: = step.::Number of Samples in Pool:: + 1'
| • | Sets the value of the Bulk Pool Volume (ul) field, based on the selected Flowcell Type: |
-exp 'if ( input.::Flowcell Type:: == ::SP:: ) {
step.::Bulk Pool Volume (ul):: = step.::Number of Flowcells to Sequence:: * 100 + 30
} ;
if ( input.::Flowcell Type:: == ::S1:: ) {
step.::Bulk Pool Volume (ul):: = step.::Number of Flowcells to Sequence:: * 100 + 30
} ;
if ( input.::Flowcell Type:: == ::S2:: ) {
step.::Bulk Pool Volume (ul):: = step.::Number of Flowcells to Sequence:: * 150 + 30
} ;
if ( input.::Flowcell Type:: == ::S4:: ) {
step.::Bulk Pool Volume (ul):: = step.::Number of Flowcells to Sequence:: * 310 + 30
} ;
| • | Calculates the Per Sample Volume (ul) to be added to the pool: |
input.::Per Sample Volume (ul):: = ( ( (input.::Final Loading Concentration (pM):: * 5 / 1000) / input.::Normalized Molarity (nM):: ) * step.::Bulk Pool Volume (ul):: ) / step.::Number of Samples in Pool::
To ensure accurate pipetting of each sample in a pool for sequencing, the Per Sample Volume (ul) value must be equal to or higher than the Minimum Per Sample Volume (ul) (default value set at 5, may be edited by the user).
Assuming the default Minimum Per Sample Volume (ul) value of 5, for a given batch:
| 1. | If the smallest Per Sample Volume (ul) value is less than 5, the LIMS automatically assigns a value of 5 to the sample's Adjusted Per Sample Volume (ul) field. |
| 2. | The LIMS then adjusts the Adjusted Per Sample Volume (ul) field value for all other samples in the batch, based on the ratio used to increase the lowest value to 5. |
| • | Calculates the Total Sample Volume (ul) field value: |
-exp 'step.::Total Sample Volume (ul):: = step.::Total Sample Volume (ul):: + input.::Adjusted Per Sample Volume (ul)::'
| • | If the Total Sample Volume is less than the Bulk Pool Volume, calculates the RSB Volume (ul) field value: |
if (step.::Total Sample Volume (ul):: >= step.::Bulk Pool Volume (ul)::) {output.::RSB Volume (ul):: = 0} else {output.::RSB Volume (ul):: = step.::Bulk Pool Volume (ul):: - step.::Total Sample Volume (ul)::} ;
| • | Copies the Flowcell Type and Loading Workflow Type values from the step inputs to the step outputs: |
output.::Flowcell Type:: = input.::Flowcell Type:: ;output.::Loading Workflow Type:: = input.::Loading Workflow Type:: ;
| • | Sets the Volume of Pool to Denature (ul) value and calculates NaOH Volume (ul) and Tris-HCl Volume (ul) values, based on the Flowcell Type: |
if ( input.::Flowcell Type:: == ::SP:: ) { output.::Volume of Pool to Denature (ul):: = 100 ; output.::NaOH Volume (ul):: = 25 ; output.::Tris-HCl Volume (ul):: = 25 } ;
if ( input.::Flowcell Type:: == ::S1:: ) { output.::Volume of Pool to Denature (ul):: = 100 ; output.::NaOH Volume (ul):: = 25 ; output.::Tris-HCl Volume (ul):: = 25 } ;
if ( input.::Flowcell Type:: == ::S2:: ) {output.::Volume of Pool to Denature (ul):: = 150 ; output.::NaOH Volume (ul):: = 37.50 ; output.::Tris-HCl Volume (ul):: = 37.50 } ;
if ( input.::Flowcell Type:: == ::S4:: ) { output.::Volume of Pool to Denature (ul):: = 310 ; output.::NaOH Volume (ul):: = 77.50 ; output.::Tris-HCl Volume (ul):: = 77.50 }
| • | Uses the NovaSeq_Standard_Bulk_Pool1.csv, NovaSeq_Standard_Bulk_Pool2.csv and NovaSeq_Standard_Bulk_Pool3.csv template files to generate a single CSV file containing information about the pool and the samples it contains. The generated file is available for download by the user on the Step Setup screen of the following step - Dilute and Denature (NovaSeq 6000 v3.0). |
&& /opt/gls/clarity/bin/java -jar /opt/gls/clarity/extensions/ngs-common/v5/EPP/DriverFileGenerator.jar -i {stepURI:v2} -u {username} -p {password} -l {compoundOutputFileLuid1} \ script:driver_file_generator \
-t /opt/gls/clarity/extensions/conf/driverfiletemplates/NovaSeq_Standard_Bulk_Pool1.csv \
-o 1.csv \ script:driver_file_generator \
-t /opt/gls/clarity/extensions/conf/driverfiletemplates/NovaSeq_Standard_Bulk_Pool2.csv \
-o 2.csv \ script:driver_file_generator \
-t /opt/gls/clarity/extensions/conf/driverfiletemplates/NovaSeq_Standard_Bulk_Pool3.csv \
-o 3.csv \
&& cat 1.csv 2.csv 3.csv > {compoundOutputFileLuid0}.csv
Set Next Step
Automatically triggered on exit of the Record Details screen, this automation sets the next step for samples to ADVANCE, advancing them to the next step in the protocol - Dilute and Denature (NovaSeq 6000 v3.0):
nextStep = ::ADVANCE::
Step UDFs/Custom Fields
There are five fields defined on the Make Bulk Pool for NovaSeq Standard (NovaSeq 6000 v3.0) step.
These are shown below in the BaseSpace Clarity LIMS v5.x Configuration area (C ustom Fields > Master Step Fields tab).
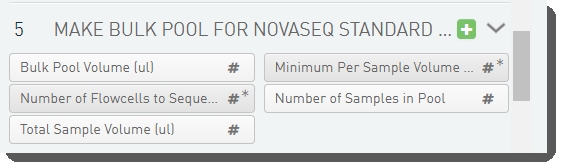
See the following table for LIMS v4.2.x and LIMS v5.x field configuration details:
|
Field Name |
Field Type |
Field Constraints/Options |
Preset Values/Additional Options and Dropdown Items |
||||||||||||
|
Bulk Pool Volume (ul) (for calculation purposes, not displayed) |
Numeric |
|
|
||||||||||||
|
Number of Flowcells to Sequence |
Numeric |
|
|
||||||||||||
|
Total Sample Volume (ul) (for calculation purposes, not displayed) |
Numeric |
|
|
||||||||||||
|
Minimum Per Sample Volume (uL) |
Numeric |
|
|
||||||||||||
|
Number of Samples in Pool (for calculation purposes, not displayed) |
Numeric |
|
|
Global UDFs/Custom Fields
The following table lists the global UDFs/custom fields that are configured to display on the Make Bulk Pool for NovaSeq Standard (NovaSeq 6000 v3.0) step.
|
Field Name |
Field Type |
Field Constraints/Options |
Preset Values/Additional Options and Dropdown Items |
|||||||||||||||||||||
|
Configured on Analyte in LIMS v4.2.x and Derived Samplein LIMS v5.x: |
||||||||||||||||||||||||
|
Flowcell Type |
Single-line Text (v4.2.x) Text Dropdown (v5.x) |
|
|
|||||||||||||||||||||
|
Loading Workflow Type |
Single-line Text (v4.2.x) Text Dropdown (v5.x) |
|
|
|||||||||||||||||||||
|
NaOH Volume (ul) |
Numeric |
|
|
|||||||||||||||||||||
|
RSB Volume (ul) |
Numeric |
|
|
|||||||||||||||||||||
|
Tris-HCl Volume (ul) |
Numeric |
|
|
|||||||||||||||||||||
|
Volume of Pool to Denature (ul) Used in Make Bulk Pool for NovaSeq Standard (NovaSeq 6000 v3.0) step only. Displays on Record Detailsscreen and in the generated CSV file. |
Numeric |
|
|
|||||||||||||||||||||
User Interaction
| 1. | On the Queue screen, user adds samples of the same Flowcell Type to the Ice Bucket and clicks View Ice Bucket. |
| 2. | On the Ice Bucket screen, user clicks Begin Work. |
| 3. | On the Pooling screen, user: |
| • | Creates a pool by dragging samples into the Pool Creator. |
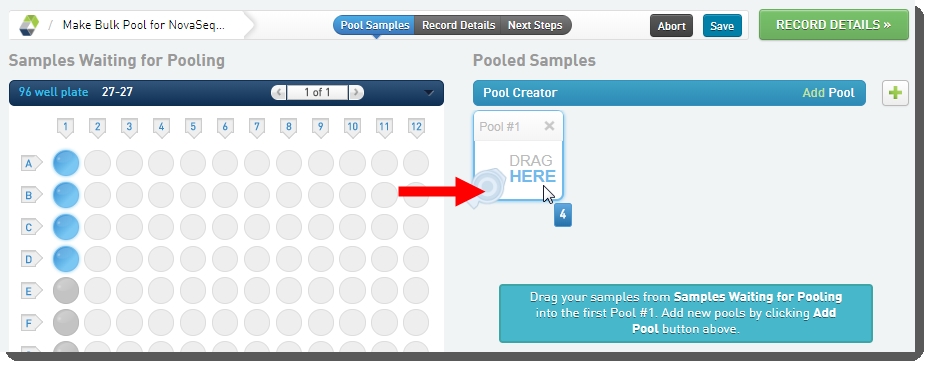
| • | Names the pool, or accepts the default name - Pool #1. |
| • | Clicks Record Details. |
On exit from the Pooling screen, the Validate Inputs Flowcell Type and Single Pool is triggered. The automation checks that all samples in the pool have been assigned the same Flowcell Type, and only one pool has been created.
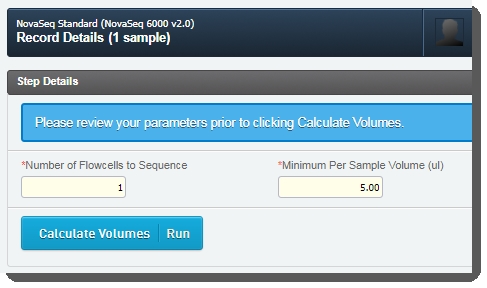
| 4. | On the Record Details screen, the Step Details area contains two required fields: |
| a. | Number of Flowcells to Sequence - The value entered in this field is used in volume calculations, to ensure that volumes are sufficient for the number of times the pool will be sequenced. |
| b. | Minimum Per Sample Volume (ul) - In many applications, and particularly where robotics are used, a minimum sample volume is required. The value in this field is used to calculate how much of each sample will be included in the pool. The field is pre-populated with the configured default value, 5 ul, but may be edited if required. |
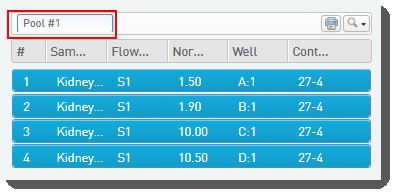
| c. | In the Sample Details table, the user can click on the pool icon to view details on the pool composition. |
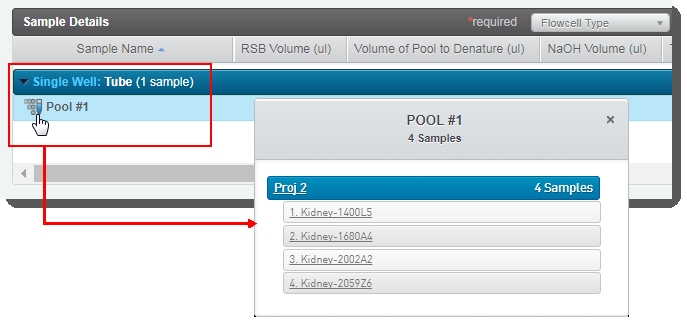
| d. | The user clicks Calculate Volumes. This triggers the Calculate Volumes automation, which performs volume calculations based on the selected Flowcell Type: |
| • | Calculates Bulk Pool Volume (ul) value. |
| • | Sets the value of the Bulk Pool Volume (ul) field, based on the selected Flowcell Type. |
| • | Calculates the Per Sample Volume (ul) value to be added to the pool. |
| • | Calculates the Total Sample Volume (ul) value. |
| • | If the Total Sample Volume is less than the Bulk Pool Volume, calculates the RSB Volume (ul) value. |
| • | Populates the Flowcell Type and Loading Workflow Type columns of the Sample Details table. |
| • | Populates the Volume of Pool to Denature (ul), NaOH Volume (ul) and T ris-HCI Volume (ul) columns of the Sample Details table. (Values are set by a script and are not editable by the user.) |
| • | Generates the Calculation File (CSV) and attaches it to the step. This file contains volume information about the pool and the individual samples it contains.The user can click on the file to download it, and then open it in Excel. |
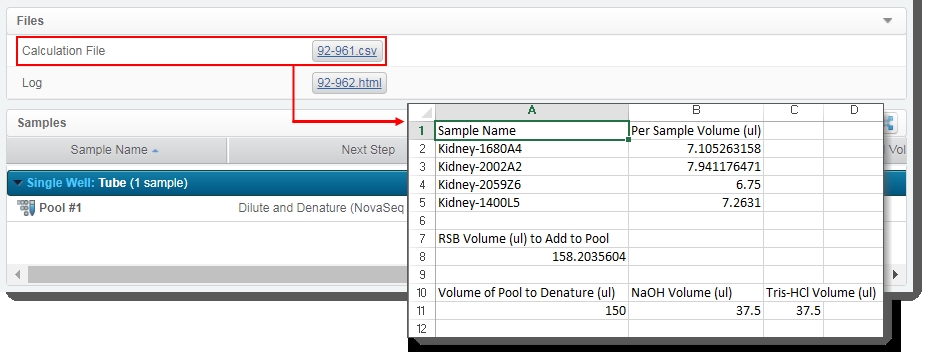
| • | The user clicks Next Steps. This triggers the Set Next Step automation. This automation sets the next step for samples to ADVANCE, advancing them to the next step in the protocol - Dilute and Denature (NovaSeq 6000 v3.0). |
| e. | On the Assign Next Steps screen: The next step is already set to Dilute and Denature (NovaSeq 6000 v3.0). |
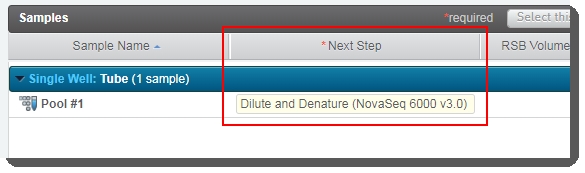
| • | User clicks Finish Step. At the end of this step, the pool of samples automatically advances to the Dilute and Denature (NovaSeq 6000 v3.0) step. |
Step 2: Dilute and Denature (NovaSeq 6000 v3.0)
In this step, pooled samples are denatured and diluted by the addition of NaOH, Tris-HCl, and Resuspension Buffer (RSB), and are manually placed by the user into the library tube that will be used in the NovaSeq run.
In addition, this step validates the run setup information and generates the sample sheet file.
Step input: Bulk pool
Step output: Library tube
The following automations are configured on the step:
| • | Routing Script |
| • | Validate Library Tube Barcode |
| • | Validate Run Setup and Generate Sample Sheet |
| • | Validate Single Input |
Dilute and Denature (NovaSeq 6000 v3.0) automation configuration - LIMS v4.2.x:
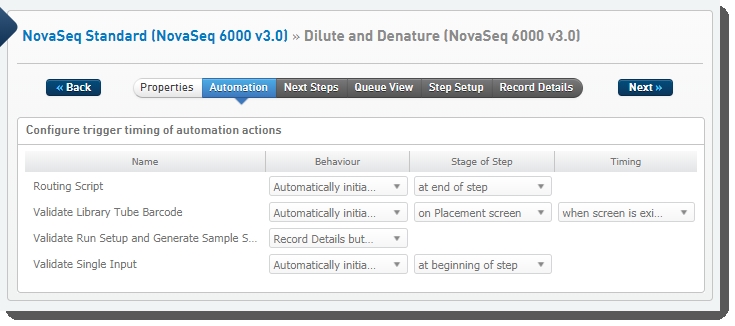
Dilute and Denature (NovaSeq 6000 v3.0) automation configuration - LIMS v5.x:
These automations are described below, in the order in which they are triggered at run time.
Validate Single Input
Automatically triggered at the beginning of the step, this automation checks that there is only one container input to the step.
script:validateSampleCount -min 1 -max 1
Validate Library Tube Barcode
Automatically triggered on exit of the Placement screen, this automation:
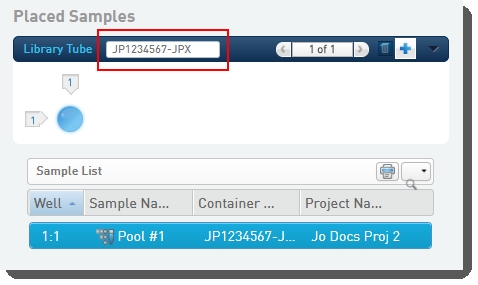
| • | Validates the library tube barcode to ensure it conforms to the barcode mask [A-Za-z]{2}[0-9]{7}-[A-Za-z]{3}: |
if (!output.container.name.matches(::[A-Za-z]{2}[0-9]{7}-[A-Za-z]{3}::)) { fail(::Invalid Library Tube Barcode. Please verify and try again.::) }
| • | Copies Flowcell Type and Loading Workflow Type field values from step inputs to outputs: |
output.::Flowcell Type:: = input.::Flowcell Type:: ; output.::Loading Workflow Type:: = input.::Loading Workflow Type::'
Validate Run Setup and Generate Sample Sheet
Automatically triggered when user clicks a button on the Record Details screen, this automation:
| • | Copies the Flowcell Type from the step input to the Run Mode field (hidden): |
step.::Run Mode:: = input.::Flowcell Type::
| • | Validates the parameters entered on the Record Details screen, which will be used to set up the run and generate the sample sheet file (see Respond to Validation/Recipe Request Call from Sequencer, below). |
| – | Experiment Name may only contain alphanumeric, dash, or underscore characters. Spaces are not permitted. |
| – | When Workflow Type = No Index, Index Read 1 must be zero. For any other Workflow Type, Index Read 1 must be greater than zero. |
if(!step.::Experiment Name::.matches(::[a-zA-Z0-9-_]+::)) {
fail(::Experiment Name contains prohibited characters. Allowed characters are: a-z, A-Z, 0-9, -, and _::)
};
if(step.::Workflow Type::== ::No Index::) {
if(step.::Index Read 1::!= 0) {
fail(::Index Read 1 must be 0 if the Workflow Type is No Index.::)
}
} else {
if(step.::Index Read 1::== 0) {
fail(::Index Read 1 must be greater than 0 if the Workflow Type is ::+ step.::Workflow Type::+ ::.::)
}
};
| • | Checks the Paired End and Read 2 Cycles field values. |
| – | If Paired End = True and Read 2 Cycles value is 0, generates an error. |
if (step.::Paired End::.toBoolean() && step.::Read 2 Cycles:: == 0) {
fail(::Read 2 Cycles must not be zero if it is Paired End read.::)
};
| • | Checks the Workflow Type and Read 2 Cycles field values. |
| – | If Workflow Type is set to Dual Index and Index Read 2 value is 0, generates an error. |
| – | If Workflow Type is set to No Index or Single Index, sets Index Read 2 to 0. |
if (step.::Workflow Type:: == ::Dual Index:: && step.::Index Read 2:: == 0) {
fail(::Index Read 2 must not be zero if Workflow Type is Dual Index.::)
};
if (step.::Workflow Type:: == ::No Index:: || step.::Workflow Type:: == ::Single Index::) {
step.::Index Read 2:: = 0
};
| • | If Paired End = False, sets Read 2 Cycles value to 0. |
if (!step.::Paired End::.toBoolean()) { step.::Read 2 Cycles:: = 0 };
| • | Checks the Flowcell Type field value. |
| – | If Flowcell Type value is not SP, checks that the value of both Read 1 Cycles and Read 2 Cycles is 151 or less. If value is greater than 151, generates an error message. |
if (input.::Flowcell Type::!=::SP:: && step.::Read 1 Cycles:: > 151) { fail(::Read 1 Cycles must not be larger than 151 if it is not SPrime Flowcell::) };if (input.::Flowcell Type::!=::SP:: && step.::Read 2 Cycles:: > 151) { fail(::Read 2 Cycles must not be larger than 151 if it is not SPrime Flowcell::)
| • | Sets the next step for samples to REMOVE: |
nextStep = ::REMOVE::
| • | Generates the sample sheet and attaches it to the step (see Sample sheet generation section, below). |
&& /opt/gls/clarity/bin/java -jar /opt/gls/clarity/extensions/ngs-common/v5/EPP/DriverFileGenerator.jar -i {stepURI:v2} -u {username} -p {password} \ script:driver_file_generator \
-t /opt/gls/clarity/extensions/conf/driverfiletemplates/NovaSeq_BCL2FASTQ_Samplesheet.csv \
-o {compoundOutputFileLuid0}.csv \
-q true \
-destLIMSID {compoundOutputFileLuid0} \
-l {compoundOutputFileLuid1}"
Routing Script
Automatically triggered on exit of the step, this automation invokes the changeWorkflow script, which routes step outputs to the NovaSeq 6000 v3.0 workflow, and queues them for the AUTOMATED - NovaSeq Run (NovaSeq 6000 v3.0) step.
Default automation command line:
bash -l -c "/opt/gls/clarity/bin/java -jar /opt/gls/clarity/extensions/ngs-common/v5/EPP/ngs-extensions.jar -u {username} -p {password} -i {stepURI:v2} -l {compoundOutputFileLuid2} script:changeWorkflow \ \
--FIELD_NAME 'N/A' \
--FIELD_VALUE 'N/A' \
--WORKFLOW 'NovaSeq 6000 v3.0' \
--STEP 'AUTOMATED - NovaSeq Run (NovaSeq 6000 v30)' \
--INPUTS_OR_OUTPUTS 'OUTPUTS'"
Step UDFs/Custom Fields
There are 16 fields defined on the Dilute and Denature (NovaSeq 6000 v3.0) step. These fields are required for sample sheet and JSON file generation.
The fields are shown below in the BaseSpace Clarity LIMS v5.x Configuration area (Custom Fields > Master Step Fields tab).
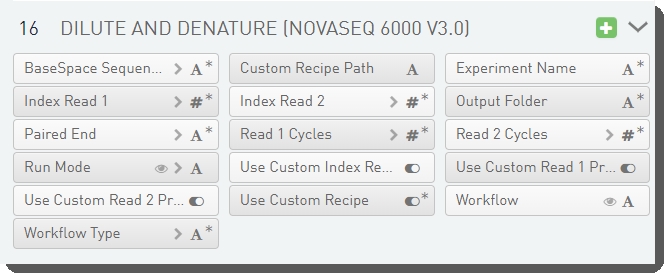
See the following table for LIMS v4.2.x and LIMS v5.x field configuration details:
|
Field Name |
Field Type |
Field Contraints/Options |
Preset Values/Additional Options and Dropdown Items |
|||||||||||||||||||||||||||
|
BaseSpace Sequence Hub Configuration |
Single-line Text (v4.2.x) Text Dropdown (v5.x) |
|
|
|||||||||||||||||||||||||||
|
Custom Recipe Path |
Single-line Text (v4.2.x) Text (v5.x) |
|
||||||||||||||||||||||||||||
|
Experiment Name |
Single-line Text (v4.2.x) Text (v5.x) |
|
||||||||||||||||||||||||||||
|
Index Read 1 |
Numeric (v4.2.x) Numeric Dropdown (v5.x) |
|
|
|||||||||||||||||||||||||||
|
Index Read 2 |
Numeric (v4.2.x) Numeric Dropdown (v5.x) |
|
|
|||||||||||||||||||||||||||
|
Output Folder |
Single-line Text (v4.2.x) Text (v5.x) |
|
||||||||||||||||||||||||||||
|
Paired End |
Single-line Text (v4.2.x) Text Dropdown (v5.x) |
|
|
|||||||||||||||||||||||||||
|
Read 1 Cycles |
Numeric (v4.2.x) Numeric Dropdown (v5.x) |
|
*Value of 251 is only supported for SPflow cell type. For all other flow cell types, maximum value is 151. |
|||||||||||||||||||||||||||
|
Read 2 Cycles |
Numeric (v4.2.x) Numeric Dropdown (v5.x) |
|
*Value of 251 is only supported for SPflow cell type. For all other flow cell types, maximum value is 151. |
|||||||||||||||||||||||||||
|
Run Mode (not displayed in user interface) |
Single-line Text (v4.2.x) Text Dropdown (v5.x) |
|
|
|||||||||||||||||||||||||||
|
Use Custom Index Read 1 Primer LIMS v5.0.5 and later: See Configuration Update for NovaSeq Integration with Clarity LIMS v5.0.5 and Later |
Check Box (v4.2.x) Toggle Switch (v5.x) |
|
|
|||||||||||||||||||||||||||
|
Use Custom Read 1 Primer LIMS v5.0.5 and later: See Configuration Update for NovaSeq Integration with Clarity LIMS v5.0.5 and Later |
Check Box (v4.2.x) Toggle Switch (v5.x) |
|
|
|||||||||||||||||||||||||||
|
Use Custom Read 2 Primer LIMS v5.0.5 and later: See Configuration Update for NovaSeq Integration with Clarity LIMS v5.0.5 and Later |
Check Box (v4.2.x) Toggle Switch (v5.x) |
|
|
|||||||||||||||||||||||||||
|
Use Custom Recipe |
Check Box (v4.2.x) Toggle Switch (v5.x) |
|
|
|||||||||||||||||||||||||||
|
Workflow |
Single-line Text (v4.2.x) Text Dropdown (v5.x) |
|
|
|||||||||||||||||||||||||||
|
Workflow Type |
Single-line Text (v4.2.x) Text Dropdown (v5.x) |
|
|
Global UDFs/Custom Fields
The following table lists the global UDFs/custom fields that are configured to display on the Dilute and Denature (NovaSeq 6000 v3.0) step.
|
Field Name |
Field Type |
Field Constraints/Options |
Preset Values/Additional Options and Dropdown Items |
|||||||||||||||||||||
|
Configured on Analyte in LIMS v4.2.x and Derived Samplein LIMS v5.x: |
||||||||||||||||||||||||
|
Flowcell Type |
Single-line Text (v4.2.x) Text Dropdown (v5.x) |
|
|
|||||||||||||||||||||
|
Loading Workflow Type |
Single-line Text (v4.2.x) Text Dropdown (v5.x) |
|
|
|||||||||||||||||||||
User Interaction
| 1. | On the Placement screen, user does the following: |
| • | Drags the pool into the library tube in the Placed Samples area. |
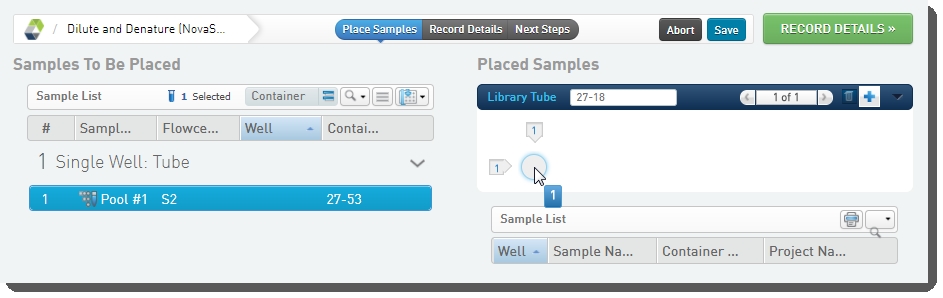
| • | Scans or types the barcode of the library tube into the Library Tube field. |
| • | Clicks Record Details. |
On exit of the Placement screen, the Validate Library Tube Barcode automation checks that the library tube barcode conforms to the barcode mask [A-Za-z]{2}[0-9]{7}-[A-Za-z]{3}, and displays an error message if this is not the case.
| 2. | On the Record Details screen: |
| • | The Reagent Lot Tracking section lets you track the NaOH, Resuspension Buffer, and Tris-HCI reagents used in the step. The user selects from the active lots displayed in each drop-down list. |
![]()
To make reagent lots available for selection in the Reagent Lot Tracking section, you must first configure and activate them. You can do this on the Reagents and Controls screen (LIMS v4.2.x) or the Configuration > Consumables screen (LIMS v5.x).
| • | The fields displayed on the Record Details screen are used to set up the run and generate the sample sheet file. |
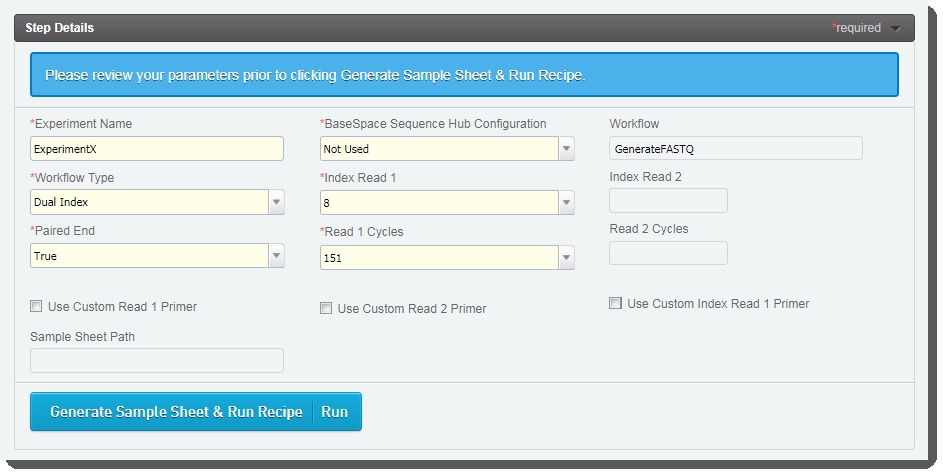
Some of these fields are auto-populated and some must be completed by the user (see the following table for details):
|
Field |
Value |
|
Experiment Name |
User enters the experiment name. Only alphanumeric characters, dashes, and underscores are permitted. Spaces are not permitted. |
|
BaseSpace Sequence Hub Configuration |
User selects from preset options: Not Used, Run Monitoring Only, Run Monitoring and Storage |
|
Workflow |
Automatically populated with preset - GenerateFASTQ |
|
Workflow Type |
User selects from preset options: No Index, Single Index, Dual Index, Custom |
|
Index Read 1 |
User selects from preset options: 0, 6, 8 - or type a value between 0 and 20 |
|
Index Read 2 |
User selects from preset options: 0, 6, 8 - or type a value between 0 and 20 |
|
Paired End |
User selects from preset options: True or False |
|
Read 1 Cycles |
User selects from preset options: 251, 151, 101, 51 - or type a value between 1 and 251 Value of 251 is only supported for SP flow cell type. For all other flow cell types, maximum value is 151. |
|
Read 2 Cycles |
User selects from preset options: 251, 151, 101, 51 - or type a value between 1 and 251 Value of 251 is only supported for SP flow cell type. For all other flow cell types, maximum value is 151. |
|
Use Custom Read 1 Primer |
User selects if applicable. |
|
Use Custom Read 2 Primer |
User selects if applicable. |
|
Use Custom Index Read 1 Primer |
User selects if applicable. |
|
Output Folder |
User enters the network path for sequencing run folder. For example: \\networkshare\run_data |
|
Use Custom Recipe |
User selects if applicable. |
|
Custom Recipe Path |
If Use Custom Recipe selected, user enters the path to the custom recipe file to be used. |
| 3. | On the Record Details screen, user clicks Validate Run Setup and Generate Sample Sheet. |
This triggers the automation script, which does the following:
| • | Validates the information provided to set up the run. |
| • | Generates the sample sheet file and attaches it to the placeholder in the Files area of the Record Detailsscreen. |
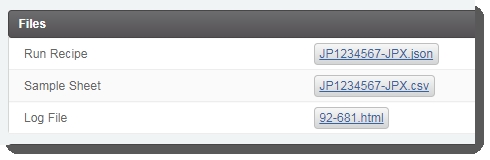
| • | Sets the value of the next step to Remove from workflow. The Routing Script automation expects this value, and requires it in order to successfully advance samples to the next step. |
| • | User clicks Next Steps. |
| 4. | On the Assign Next Steps screen: The Next Step field for all samples is pre-populated with Remove from workflow. |
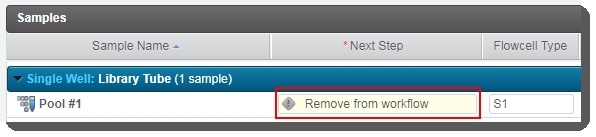
The user must not change this value. If Next Step is not set to Remove from workflow, the routing script will not be able to route samples correctly.
| • | User clicks Finish Step. |
| 5. | On exit from the step: |
| • | The Routing Script automation is triggered and samples are routed to the AUTOMATED - NovaSeq Run (NovaSeq 6000 v3.0) protocol. |
| • | In Lab View, the pool of samples is queued for the AUTOMATED - NovaSeq Run (NovaSeq 6000 v3.0) step. |
At this point in the workflow, the user interaction ends. The integration automatically starts the AUTOMATED - NovaSeq Run (NovaSeq 6000 v3.0) step and data from the run is parsed back into the LIMS.
Protocol 3: NovaSeq Xp (NovaSeq 6000 v3.0)
Samples are routed to this protocol if their Loading Workflow Type value is set to NovaSeq Xp.
Samples are pooled and added to lanes on the NovaSeq flow cell type selected in the Define Run Format (NovaSeq 6000 v3.0) step. At the end of this protocol, the flow cell is sent to the AUTOMATED - NovaSeq Run (NovaSeq 6000 v3.0) protocol.
This protocol contains three steps, described below:
| • | Step 1: Make Bulk Pool for NovaSeq Xp (NovaSeq 6000 v3.0) |
| • | Step 2: Dilute, Denature and ExAmp (NovaSeq 6000 v3.0) |
| • | Step 3: Load to Flowcell (NovaSeq 6000 v3.0) |
Step 1: Make Bulk Pool for NovaSeq Xp (NovaSeq 6000 v3.0)
In this step, user manually places libraries into a pool.
User must only create one pool per step.
Step input: NTP (normalized libraries)
Step output: Bulk pool
The following automations are configured on the step:
| • | Calculate Volumes |
| • | Set Next Step |
| • | Validate Inputs Flowcell Type and Single Pool |
| • | Validate Unique Indexes (Not used. This functionality is handled by the core LIMS configuration for pooling.) |
Make Bulk Pool for NovaSeq Xp (NovaSeq 6000 v3.0) automation configuration - LIMS v4.2.x:
Make Bulk Pool for NovaSeq Xp (NovaSeq 6000 v3.0) automation configuration - LIMS v5.x:
These automations are described below, in the order in which they are triggered at run time.
Validate Inputs Flowcell Type and Single Pool
Automatically triggered on exit of the Pooling screen, this automation:
| • | Checks that all samples in the pool have the same Flowcell Type assigned to them. |
| • | Checks that only one pool has been created. |
| • | Changes the value of error and logging messages to reference the type of pool passed as 'bulk' pool. |
/opt/gls/clarity/bin/java \-jar /opt/gls/clarity/extensions/ngs-common/v5/EPP/ngs-extensions.jar \script:validate_flowcell_for_input_pools \
-i {stepURI:v2} \
-u {username} \
-p {password} \
-l {compoundOutputFileLuid1} \
-validateSingleOutput true \
-poolType bulk"
Calculate Volumes
Automatically triggered when the user clicks the Calculate Volumes button on the Record Details screen, this automation:
| • | Calculates the number of samples in the pool: |
step.::Number of Samples in Pool:: = step.::Number of Samples in Pool:: + 1
| • | Sets the value of the Bulk Pool Volume (ul) field,based on the selected Flowcell Type: |
if ( input.::Flowcell Type:: == ::SP:: ) {
step.::Bulk Pool Volume (ul):: = step.::Number of Lanes to Sequence:: * 18 + 30
} ;
if ( input.::Flowcell Type:: == ::S1:: ) {
step.::Bulk Pool Volume (ul):: = step.::Number of Lanes to Sequence:: * 18 + 30
} ;
if ( input.::Flowcell Type:: == ::S2:: ) {
step.::Bulk Pool Volume (ul):: = step.::Number of Lanes to Sequence:: * 22 + 30
} ;
if ( input.::Flowcell Type:: == ::S4:: ) {
step.::Bulk Pool Volume (ul):: = step.::Number of Lanes to Sequence:: * 30 + 30
} ;
| • | Calculates the Per Sample Volume (ul) to be added to the pool: |
input.::Per Sample Volume (ul)::= (((input.::Final Loading Concentration (pM)::* 5/ 1000) / input.::Normalized Molarity (nM)::) * step.::Bulk Pool Volume (ul)::) / step.::Number of Samples in Pool::
To ensure accurate pipetting of each sample in a pool for sequencing, the Per Sample Volume (ul) value must be equal to or higher than the Minimum Per Sample Volume (ul) (default value set at5, may be edited by the user).
Assuming the default Minimum Per Sample Volume (ul) value of 5, for a given batch:
| 1. | If the smallest Per Sample Volume (ul) value is less than 5, the LIMS automatically assigns a value of 5 to the sample's Adjusted Per Sample Volume (ul) field. |
| 2. | The LIMS then adjusts the Adjusted Per Sample Volume (ul) field value for all other samples in the batch, based on the ratio used to increase the lowest value to 5. |
| • | Calculates the Total Sample Volume (ul) field value: |
step.::Total Sample Volume (ul):: = step.::Total Sample Volume (ul):: + input.::Adjusted Per Sample Volume (ul)::
| • | If the Total Sample Volume is less than the Bulk Pool Volum e, calculates the RSB Volume (ul) field value: |
if (step.::Total Sample Volume (ul):: >= step.::Bulk Pool Volume (ul)::) {output.::RSB Volume (ul):: = 0} else {output.::RSB Volume (ul):: = step.::Bulk Pool Volume (ul):: - step.::Total Sample Volume (ul)::} ;
| • | Copies the Flowcell Type and Loading Workflow Type values from the step inputs to the step outputs: |
output.::Flowcell Type::= input.::Flowcell Type::;output.::Loading Workflow Type::= input.::Loading Workflow Type::;
| • | Uses the NovaSeq_Xp_Bulk_Pool.csv and NovaSeq_Xp_Bulk_Pool2.csv template files to generate a single CSV file containing information about the bulk pool and the samples it contains. The generated file is available for download by the user on the Step Setup screen of the following step - Dilute, Denature and ExAmp (NovaSeq 6000 v3.0). |
&& /opt/gls/clarity/bin/java \-jar /opt/gls/clarity/extensions/ngs-common/v5/EPP/DriverFileGenerator.jar \
-i {stepURI:v2} \
-u {username} \
-p {password} \
-l {compoundOutputFileLuid1} \ script:driver_file_generator \
-t /opt/gls/clarity/extensions/conf/driverfiletemplates/NovaSeq_Xp_Bulk_Pool.csv \
-o {compoundOutputFileLuid0}.csv \ script:driver_file_generator \
-t /opt/gls/clarity/extensions/conf/driverfiletemplates/NovaSeq_Xp_Bulk_Pool2.csv \
-o append.csv \
&& cat append.csv >> {compoundOutputFileLuid0}.csv"
Set Next Step
Automatically triggered on exit of the Record Details screen, this automation:
| • | Copies the Flowcell Type values from the step inputs to the step outputs. |
| • | Sets the next step for samples to ADVANCE, advancing them to the next step in the protocol: |
output.::Flowcell Type:: = input.::Flowcell Type:: ; nextStep = ::ADVANCE::
Step UDFs/Custom Fields
There are five fields defined on the Make Bulk Pool for NovaSeq Xp (NovaSeq 6000 v3.0) step.
These are shown below in the BaseSpace Clarity LIMS v5.x Configuration area (Custom Fields > Master Step Fields tab).
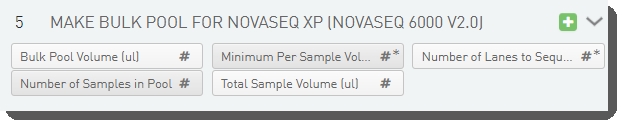
See the following table for LIMS v4.2.x and LIMS v5.x field configuration details:
|
Field Name |
Field Type |
Field Constraints/Options |
Preset Values/Additional Options and Dropdown Items |
||||||||||||
|
Bulk Pool Volume (ul) (for calculation purposes, not displayed) |
Numeric |
|
|
||||||||||||
|
Number of Lanes to Sequence |
Numeric |
|
|
||||||||||||
|
Total Sample Volume (ul) (for calculation purposes, not displayed) |
Numeric |
|
|
||||||||||||
|
Minimum Per Sample Volume (uL) |
Numeric |
|
|
||||||||||||
|
Number of Samples in Pool (for calculation purposes, not displayed) |
Numeric |
|
|
Global UDFs/Custom Fields
The following table lists the global UDFs/custom fields that are configured to display on the Make Bulk Pool for NovaSeq Xp (NovaSeq 6000 v3.0) step:
|
Field Name |
Field Type |
Field Constraints/Options |
Preset Values/Additional Options and Dropdown Items |
|||||||||||||||||||||
|
Configured on Analyte in LIMS v4.2.x and Derived Samplein LIMS v5.x: |
||||||||||||||||||||||||
|
RSB Volume (ul) |
Numeric |
|
|
|||||||||||||||||||||
|
Flowcell Type |
Single-line Text (v4.2.x) Text Dropdown (v5.x) |
|
|
|||||||||||||||||||||
|
Loading Workflow Type |
Single-line Text (v4.2.x) Text Dropdown (v5.x) |
|
|
|||||||||||||||||||||
User Interaction
| 1. | On the Queue screen, user adds samples of the same Flowcell Type to the Ice Bucket and clicks View Ice Bucket. |
| 2. | On the Ice Bucket screen, user clicks Begin Work. |
| 3. | On the Pooling screen, user: |
| • | Drags samples over into the Pool Creator to create a pool. |

| • | Names the pool, or accepts the default name - Pool #1. |
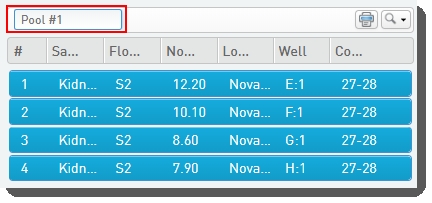
| • | Clicks Record Details. |
On exit from the Pooling screen, the Validate Inputs Flowcell Type and Single Pool is triggered. The automation checks that all samples in the pool have been assigned the same Flowcell Type, and only one pool has been created.
| 4. | On the Record Details screen, the Step Details area contains two required fields: |
| • | Number of Lanes to Sequence: The value entered in this field is used in volume calculations, to ensure that volumes are sufficient for the number of times the pool will be sequenced. |
| • | Minimum Per Sample Volume (ul): In many applications, and particularly where robotics are used, a minimum sample volume is required. The value in this field is used to calculate how much of each sample will be included in the pool. |
The field is pre-populated with the configured default value, 5 ul, but may be edited if required.
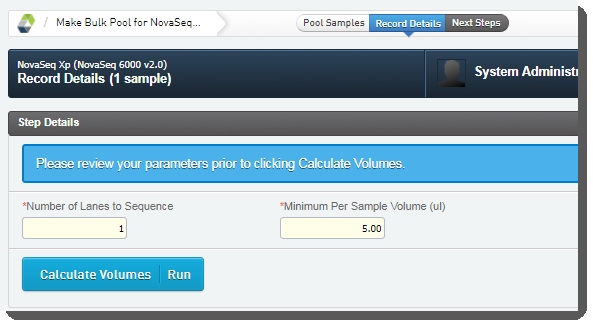
| • | In the Sample Details table, the user can click on the pool icon to view details on the pool composition. |

| • | The user clicks Calculate Volumes. This triggers the Calculate Volumes automation, which performs volume calculations based on the selected Flowcell Type: Calculates Bulk Pool Volume (ul) value. |
| – | Calculates the Per Sample Volume (ul) value to be added to the pool. |
| – | Calculates the Total Sample Volume (ul) value. |
| – | If the Total Sample Volume is less than the Bulk Pool Volume, calculates the RSB Volume (ul) value. |
| – | Populates the Flowcell Type and Loading Workflow Type columns of the Sample Details table. |
| – | Generates the Calculation File (CSV) and attaches it to the step. This file contains volume information about the samples and RSB buffer to add to the pool.The user can click on the file to download it, and then open it in Excel. |
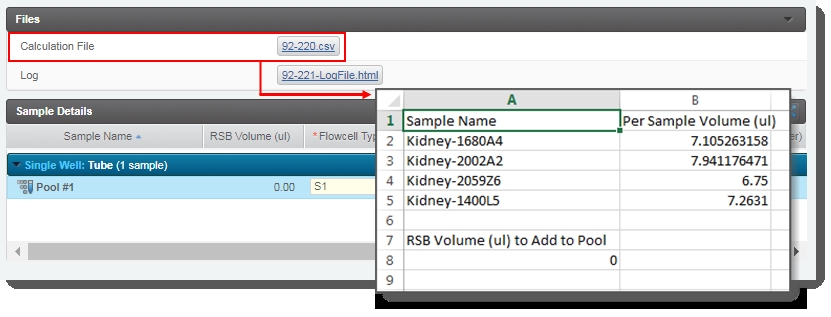
| – | The user clicks Next Steps. This triggers the Set Next Step automation. This automation sets the next step for samples to ADVANCE, advancing them to the next step in the protocol - Dilute, Denature and ExAmp (NovaSeq 6000 v3.0). |
| 5. | On the Assign Next Steps screen: |
| • | The next step is already set to Dilute, Denature and ExAmp (NovaSeq 6000 v3.0). |
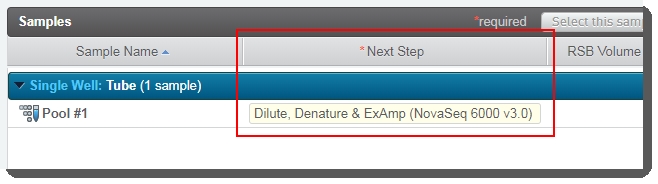
| • | User clicks Finish Step. At the end of this step, the pool of samples automatically advances to the Dilute, Denature and ExAmp (NovaSeq 6000 v3.0) step. |
Step 2: Dilute, Denature and ExAmp (NovaSeq 6000 v3.0)
In this step, pooled samples are denatured and diluted by the addition of DPX, NaOH, Tris-HCl, and RSB. The user manually creates working pools based on the number of lanes they want to sequence.
Step input: Bulk pool
Step output: Working pool - variable number, the user chooses how many working pools to create per bulk pool
The following automations are configured on the step:
| • | Calculate Volumes |
| • | Set Next Step |
| • | Validate Inputs Flowcell Type |
Dilute, Denature and ExAmp (NovaSeq 6000 v3.0) automation configuration - LIMS v4.2.x:
Dilute, Denature and ExAmp (NovaSeq 6000 v3.0) automation configuration - LIMS v5.x:
These automations are described below, in the order in which they are triggered at run time.
Validate Inputs Flowcell Type
Automatically triggered at the beginning of the step, this automation invokes the validate_flowcell_for_input_pools script. This script does the following:
| • | Checks the inputs to the step and validates that the Flowcell Type field has been set to a valid value (SP, S1, S2, or S4), and that all inputs have the same value for this field. |
| • | The container type selected matches the value in the Flowcell Type field. |
| • | Validates that the number of outputs matches the number of lanes on the selected flow cell type. If validation fails, an error message informs the user that the number of working pools does not match the number of lanes available on the flow cell. |
bash -l -c "/opt/gls/clarity/bin/java \
-jar /opt/gls/clarity/extensions/ngs-common/v5/EPP/ngs-extensions.jar \script:validate_flowcell_for_input_pools \
-i {stepURI:v2} \
-u {username} \
-p {password} \
-l {compoundOutputFileLuid1}"
Calculate Volumes
Automatically triggered on entry to of the Record Details screen, this automation sets the value of the following fields, based on the Flowcell Type:
| • | BP Aliquot Volume (ul) |
| • | NaOH Volume (ul) |
| • | Tris-HCl Volume (ul) |
| • | DPX1 Volume (ul) |
| • | DPX2 Volume (ul) |
| • | DPX3 Volume (ul) |
| • | Mastermix per Lane (ul) |
'if ( input.::Flowcell Type:: == ::SP:: ) {
output.::BP Aliquot Volume (ul):: = 18 ;
output.::NaOH Volume (ul):: = 4.50 ;
output.::Tris-HCl Volume (ul):: = 4.50 ;
step.::DPX1 Volume (ul):: = 126 ;
step.::DPX2 Volume (ul):: = 18 ;
step.::DPX3 Volume (ul):: = 66 ;
output.::Mastermix per Lane (ul):: = 63};
if (input.::Flowcell Type:: == ::S1::) {
output.::BP Aliquot Volume (ul):: = 18;
output.::NaOH Volume (ul):: = 4.50;
output.::Tris-HCl Volume (ul):: = 4.50;
step.::DPX1 Volume (ul):: = 126;
step.::DPX2 Volume (ul):: = 18;
step.::DPX3 Volume (ul):: = 66;
output.::Mastermix per Lane (ul):: = 63};
if (input.::Flowcell Type:: == ::S2::) {
output.::BP Aliquot Volume (ul):: = 22;
output.::NaOH Volume (ul):: = 5.50;
output.::Tris-HCl Volume (ul):: = 5.50;
step.::DPX1 Volume (ul):: = 126;
step.::DPX2 Volume (ul):: = 18;
step.::DPX3 Volume (ul):: = 66;
output.::Mastermix per Lane (ul):: = 77};
if (input.::Flowcell Type:: == ::S4::) {
output.::BP Aliquot Volume (ul):: = 30;
output.::NaOH Volume (ul):: = 7.50;
output.::Tris-HCl Volume (ul):: = 7.50;
step.::DPX1 Volume (ul):: = 315;
step.::DPX2 Volume (ul):: = 45;
step.::DPX3 Volume (ul):: = 165;
output.::Mastermix per Lane (ul):: = 105
};
The automation also:
| • | Copies the Flowcell Type and Loading Workflow Type values from the step inputs to the step outputs: |
output.::Flowcell Type::= input.::Flowcell Type::;
output.::Loading Workflow Type::= input.::Loading Workflow Type::;
| • | Uses the NovaSeq_Xp_Working_Pool.csv and NovaSeq_Xp_Working_Pool2.csv template files to generate a single CSV file containing information about the DPX volume, and the volume of BP Aliquot, Mastermix, NaOH, and Tris-HCI to add per working pool. The generated file is available for download by the user on the Step Setup screen of the following step - Load to Flowcell (NovaSeq 6000 v3.0). |
and /opt/gls/clarity/bin/java \
-jar /opt/gls/clarity/extensions/ngs-common/v5/EPP/DriverFileGenerator.jar \
-i {stepURI:v2} \
-u {username} \
-p {password} \
-l {compoundOutputFileLuid1} \script:driver_file_generator \
-t /opt/gls/clarity/extensions/conf/driverfiletemplates/NovaSeq_Xp_Working_Pool.csv \
-o {compoundOutputFileLuid0}.csv \script:driver_file_generator \
-t /opt/gls/clarity/extensions/conf/driverfiletemplates/NovaSeq_Xp_Working_Pool2.csv \
-o append.csv \
&& cat append.csv >> {compoundOutputFileLuid0}.csv"
Set Next Step
Automatically triggered on exit of the Record Details screen, this automation:
| • | Copies the Flowcell Type values from the step inputs to the step outputs. |
| • | Sets the next step for samples to ADVANCE, advancing them to the next step in the protocol: |
output.::Flowcell Type:: = input.::Flowcell Type:: ; nextStep = ::ADVANCE::
Step UDFs/custom fields
There are 3 fields defined on the Dilute, Denature and ExAmp (NovaSeq 6000 v3.0) step. (These field values are set by a script and are not editable by the user running the step.)
The fields are shown below in the BaseSpace Clarity LIMS v5.x Configuration area (Custom Fields > Master Step Fields tab).
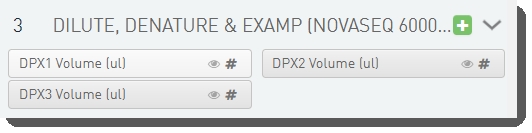
See the following table for LIMS v4.2.x and LIMS v5.x field configuration details:
|
Field Name |
Field Type |
Field Constraints/Options |
Preset Values/Additional Options and Dropdown Items |
|||||||||
|
DPX1 Volume (ul) |
Numeric |
|
|
|||||||||
|
DPX2 Volume (ul) |
Numeric |
|
|
|||||||||
|
DPX3 Volume (ul) |
Numeric |
|
|
Global UDFs/Custom Fields
The following table lists the global UDFs/custom fields that are configured to display on the Dilute, Denature and ExAmp (NovaSeq 6000 v3.0) step:
|
Field Name |
Field Type |
Field Constraints/Options |
Preset Values/Additional Options and Dropdown Items |
|||||||||||||||||||||
|
Configured on Analyte in LIMS v4.2.x and Derived Sample in LIMS v5.x: |
||||||||||||||||||||||||
|
BP Aliquot Volume (ul) |
Numeric |
|
|
|||||||||||||||||||||
|
Flowcell Type |
Single-line Text (v4.2.x) Text Dropdown (v5.x) |
|
|
|||||||||||||||||||||
|
Loading Workflow Type |
Single-line Text (v4.2.x) Text Dropdown (v5.x) |
|
|
|||||||||||||||||||||
|
Mastermix per Lane (ul) |
Numeric |
|
|
|||||||||||||||||||||
|
NaOH Volume (ul) |
Numeric |
|
|
|||||||||||||||||||||
|
Tris-HCl Volume (ul) |
Numeric |
|
|
|||||||||||||||||||||
User Interaction
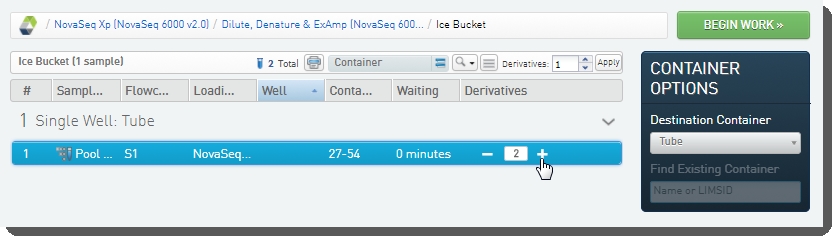
| 1. | On the Queue screen, user adds the pool to the Ice Bucket and clicks View Ice Bucket. |
| 2. | On the Ice Bucket screen, user (optionally) sets the number of derivatives to create - these will be placed into the flow cell lanes, and clicks Begin Work. |
On entry to the Record Details screen, the Calculate Volumes automation is triggered. This automation sets the following values based on the selected Flowcell Type (calculation details are provided inCalculate Volumes automation section, above):
| • | BP Aliquot Volume (ul) |
| • | NaOH Volume (ul) |
| • | Tris-HCl Volume (ul) |
| • | DPX1 Volume (ul) |
| • | DPX2 Volume (ul) |
| • | DPX3 Volume (ul) |
| • | Mastermix per Lane (ul) |
| • | Populates the Flowcell Type and Loading Workflow Type columns of the Sample Details table. |
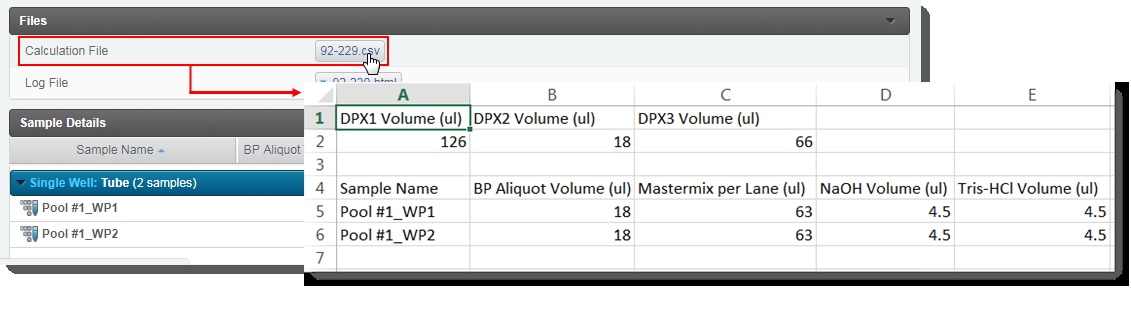
| • | Generates the Calculation File (CSV) and attaches it to the step. This file contains information about the DPX Mastermix volume, and the volume of Mastermix, NaOH, and Tris-HCI to add per working pool. The user can click on the file to download it and then open it in Excel. |
| 3. | On the Record Details screen: The Reagent Lot Tracking section lets you track the DPX1, DPX2, DPX3, NaOH, RSB, and Tris-HCI reagent lots. The user selects from the active lots displayed in each drop-down list. |
![]()
To make reagent lots available for selection in the Reagent Lot Tracking section, you must first configure and activate them. You can do this on the Reagents and Controls screen (LIMS v4.2.x) or the Configuration > Consumables screen (LIMS v5.x).
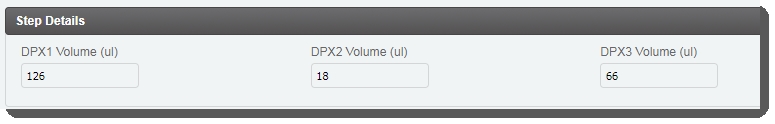
| • | In the Step Details area, the DPX1, DPX2, and DPX3 reagent volume values are already populated. (These values are set by a script and are not editable by the user running the step.) |
| • | In the Sample Details table: |
| – | The user can click on the pool icon to view details on the working pool composition. |
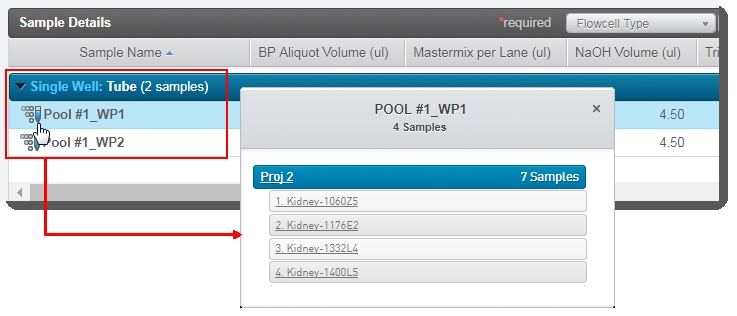
| – | The BP Aliquot, Mastermix per lane, NaOH, and Tris-HCI volume values for each working pool are already populated. (These values are set by a script and are not editable by the user running the step.) |
| – | The working pool number is appended to the bulk pool name. This allows users to quickly identify which working pools are derived from the same bulk pool. |
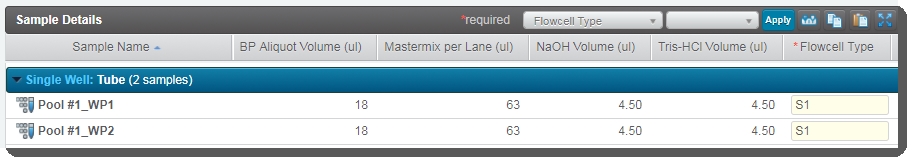
| • | In the Files area, the user can click on the Calculation File (CSV) file to download it and then open it in Excel to view information about the DPX Mastermix volume, and the volume of Mastermix, NaOH. and Tris-HCI to add per working pool. |
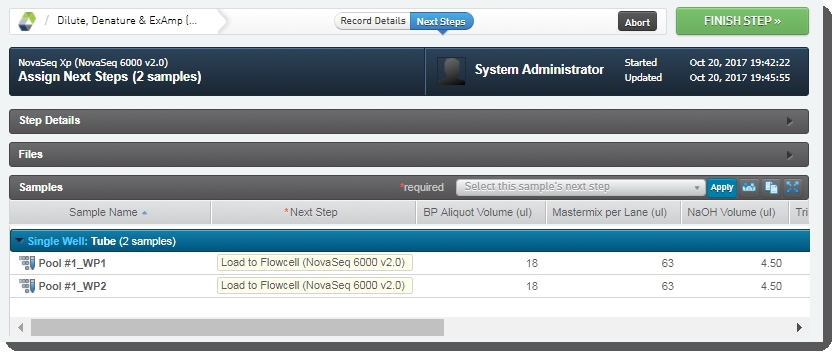
| • | User clicks Next Steps, which triggers the Set Next Step automation (described above). |
| 4. | On the Assign Next Steps screen, the next step is already set to Load to Flowcell (NovaSeq 6000 v3.0). User clicks Finish Step. |
Step 3: Load to Flowcell (NovaSeq 6000 v3.0)
In this step, the user scans the flow cell barcode into the LIMS, and then manually places the working pools into the lanes of the flow cell that will be used in the NovaSeq run. In addition, this step validates the run setup information and generates the sample sheet file.
Step input: Working pool
Step output: Flow cell (output containers: SP, S1, and S2 with 2 lanes, and S4 with 4 lanes)
The following automations are configured on the step:
| • | Validate Run Setup and Generate Sample Sheet |
| • | Validate Flowcell Barcode |
| • | Validate Inputs and Selected Container |
Load to Flowcell (NovaSeq 6000 v3.0) automation configuration - LIMS v4.2.x:
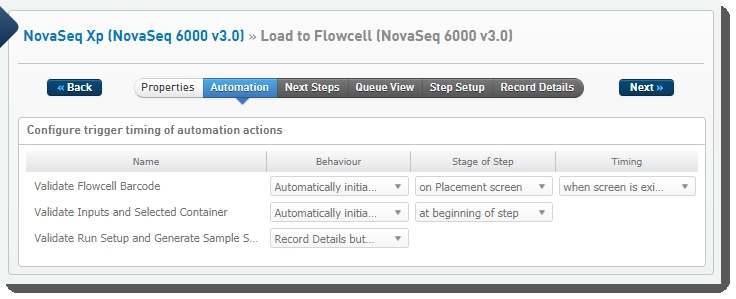
Load to Flowcell (NovaSeq 6000 v3.0) automation configuration - LIMS v5.x:
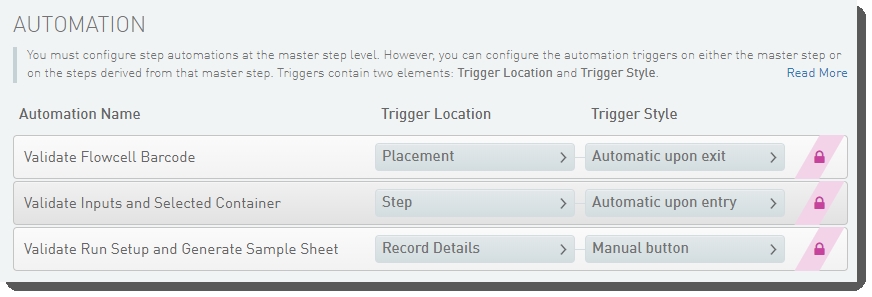
These automations are described below, in the order in which they are triggered at run time.
Validate Inputs and Selected Container
Automatically triggered at the beginning of the step, this automation invokes the validate_flowcell_for_input_pools and validateSelectedContainer scripts. These scripts validate the step inputs and the container selected by the user, as follows:
| • | Check that the Flowcell Type field has been set to a valid value (SP, S1, S2, or S4), and that all inputs have the same value. |
| • | Check that the number of outputs matches the number of lanes on the selected flow cell type. If validation fails, an error message informs the user that the number of working pools does not match the number of lanes available on the flow cell. |
The following table defines the number of samples allowed:
|
Flow Cell Type |
Number of Working Pools |
|
SP |
2 |
|
S1 |
2 |
|
S2 |
2 |
|
S4 |
4 |
| • | Check that the container type selected on entry to the Placement screen matches the value in the Flowcell Type field. If validation fails, displays an error message. |
bash -l -c "/opt/gls/clarity/bin/java \
-jar /opt/gls/clarity/extensions/ngs-common/v5/EPP/ngs-extensions.jar \script:validate_flowcell_for_input_pools \
-i {stepURI:v2} \
-u {username} \
-p {password} \
-l {compoundOutputFileLuid1} \
-validateSelectedContainer true"
Validate Flowcell Barcode
Automatically triggered on exit of the Placement screen, this automation:
| • | Validates the flow cell barcode scanned into the LIMS by the user, using the following logic: |
if (input.::Flowcell Type:: == ::SP:: && !output.container.name.matches( ::[A-Za-z0-9]{5}DRXX:: ) ) { \
fail ( ::Invalid Flowcell Barcode. Please verify and try again.:: ) \
} ; \
if (input.::Flowcell Type:: == ::S1:: && !output.container.name.matches( ::[A-Za-z0-9]{5}DRXX:: ) ) { \
fail ( ::Invalid Flowcell Barcode. Please verify and try again.:: ) \
} ; \
if (input.::Flowcell Type:: == ::S2:: && !output.container.name.matches( ::[A-Za-z0-9]{5}DMXX:: ) ) { \
fail ( ::Invalid Flowcell Barcode. Please verify and try again.:: ) \
} ; \
if (input.::Flowcell Type:: == ::S4:: && !output.container.name.matches( ::[A-Za-z0-9]{5}DSXX:: ) ) { \
fail ( ::Invalid Flowcell Barcode. Please verify and try again.:: ) \
} ;
| • | Copies the Flowcell Type and Loading Workflow Type field values from step inputs to outputs: |
output.::Flowcell Type:: = input.::Flowcell Type:: ; \
output.::Loading Workflow Type:: = input.::Loading Workflow Type::'
Validate Run Setup and Generate Sample Sheet
Automatically triggered when user clicks a button on the Record Details screen, this automation:
| • | Copies the Flowcell Type from the step input to the Run Mode field (hidden): |
step.::Run Mode:: = input.::Flowcell Type::
| • | Validates the parameters entered on the Record Details screen, which will be used to set up the run and generate the sample sheet file. (See Respond to Validation/Recipe Request Call from Sequencer section, below.) |
| – | Experiment Name may only contain alphanumeric, dash, or underscore characters. Spaces are not permitted. |
| – | When Workflow Type = No Index, Index Read 1 must be zero. For any other Workflow Type, Index Read 1 must be greater than zero. |
if(!step.::Experiment Name::.matches(::[a-zA-Z0-9-_]+::)) { \
fail(::Experiment Name contains prohibited characters. Allowed characters are: a-z, A-Z, 0-9, -, and _::) \
}; \
if(step.::Workflow Type::== ::No Index::) { \
if(step.::Index Read 1::!= 0) { \
fail(::Index Read 1 must be 0 if the Workflow Type is No Index.::) \
} \
} else { \
if(step.::Index Read 1::== 0) { \
fail(::Index Read 1 must be greater than 0 if the Workflow Type is ::+ step.::Workflow Type::+ ::.::) \
} \
};
| • | Checks the Paired End and Read 2 Cycles field values. |
| – | If Paired End = False, sets Read 2 Cycles value to 0. |
| – | If Paired End= True and 2 Cycles value is 0, generates an error. |
if (!step.::Paired End::.toBoolean()) {
step.::Read 2 Cycles:: = 0 \
}; \
if (step.::Paired End::.toBoolean() && step.::Read 2 Cycles:: == 0) \{ \
fail(::Read 2 Cycles must not be zero if it is Paired End read.::) \
};
| • | Checks the Workflow Type and Read 2 Cycles field values. |
| – | If Workflow Type is set to Dual Index and Index Read 2 value is 0, generates an error. |
| – | If Workflow Type is set to No Index or Single Index, sets Index Read 2 to 0. |
if (step.::Workflow Type:: == ::Dual Index:: && step.::Index Read 2:: == 0) {
fail(::Index Read 2 must not be zero if Workflow Type is Dual Index.::)
};
if (step.::Workflow Type:: == ::No Index:: || step.::Workflow Type:: == ::Single Index::) {
step.::Index Read 2:: = 0
};
| • | Copies the Flowcell Type value from the step inputs to the step outputs: |
output.::Flowcell Type::= input.::Flowcell Type::;
| • | Sets the next step for samples to ADVANCE, advancing them to the next step in the protocol - AUTOMATED - Run (NovaSeq 6000) v3.0. |
nextStep = ::ADVANCE::
| • | Generates the sample sheet and attaches it to the step (see Sample Sheet Generation, below). |
&& /opt/gls/clarity/bin/java \
-jar /opt/gls/clarity/extensions/ngs-common/v5/EPP/DriverFileGenerator.jar \
-i {stepURI:v2} \
-u {username} \
-p {password} \
script:driver_file_generator \
-t /opt/gls/clarity/extensions/conf/driverfiletemplates/NovaSeq_BCL2FASTQ_Samplesheet.csv \
-o {compoundOutputFileLuid0}.csv \
-q true \
-destLIMSID {compoundOutputFileLuid0} \
-l {compoundOutputFileLuid1}"
Step UDFs/Custom Fields
There are 17 fields defined on the Load to Flowcell (NovaSeq 6000 v3.0) step.
These are shown below in the BaseSpace Clarity LIMS v5.x Configuration area (Custom Fields > Master Step Fields tab).
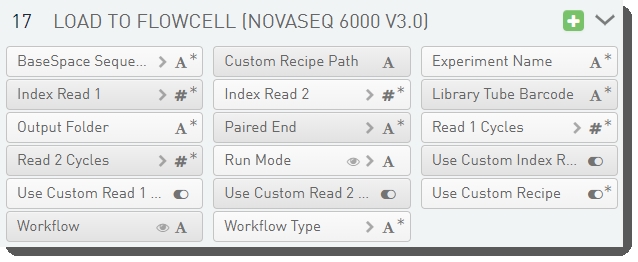
See the following table for LIMS v4.2.x and LIMS v5.x field configuration details:
|
Field Name |
Field Type |
Field Contraints/Options |
Preset Values/Additional Options and Dropdown Items |
|||||||||||||||||||||
|
BaseSpace Sequence Hub Configuration |
Check Box (v4.2.x) Toggle Switch (v5.x) |
|
|
|||||||||||||||||||||
|
Use Custom Recipe |
Check Box (v4.2.x) Toggle Switch (v5.x) |
|
|
|||||||||||||||||||||
|
Workflow |
Single-line Text (v4.2.x) Text (v5.x) |
|
|
|||||||||||||||||||||
|
Workflow Type |
Single-line Text (v4.2.x) Text Dropdown (v5.x) |
|
|
Global UDFs/Custom Fields
The following table lists the global UDFs/custom fields that are configured to display on the Load to Flowcell (NovaSeq 6000 v3.0) step.
|
Field Name |
Field Type |
Field Constraints/Options |
Preset Values/Additional Options and Dropdown Items |
|||||||||||||||||||||
|
Configured on Analyte in LIMS v4.2.x and Derived Sample in LIMS v5.x: |
||||||||||||||||||||||||
|
Flowcell Type |
Single-line Text (v4.2.x) Text Dropdown (v5.x) |
|
|
|||||||||||||||||||||
|
Loading Workflow Type |
Single-line Text (v4.2.x) Text Dropdown (v5.x) |
|
|
|||||||||||||||||||||
User Interaction
| 1. | On the Ice Bucket screen, user: |
| • | Selects the appropriate flow cell type from the Destination Container drop-down list in the Container Options panel. |
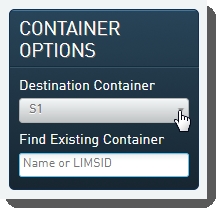
| • | Clicks Begin Work. |
| 2. | On the Placement screen, user: |
| • | Drags the pool(s) from the left of the screen over into the Placed Samples area on the right. |
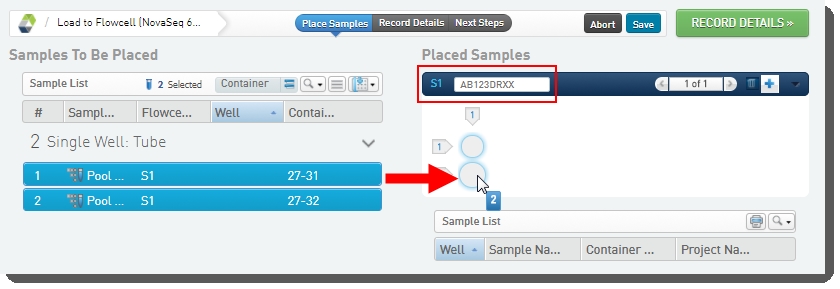
| • | Scans or types the barcode of the flow cell into the flow cell field. |
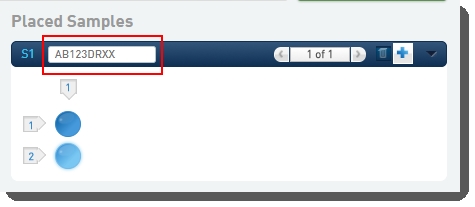
| • | Clicks Record Details. |
On exit of the Placement screen, the Validate Flowcell Barcode automation validates the container barcode (described above).
| 3. | The fields displayed on the Record Details screen are used to set up the run and generate the sample sheet file. |
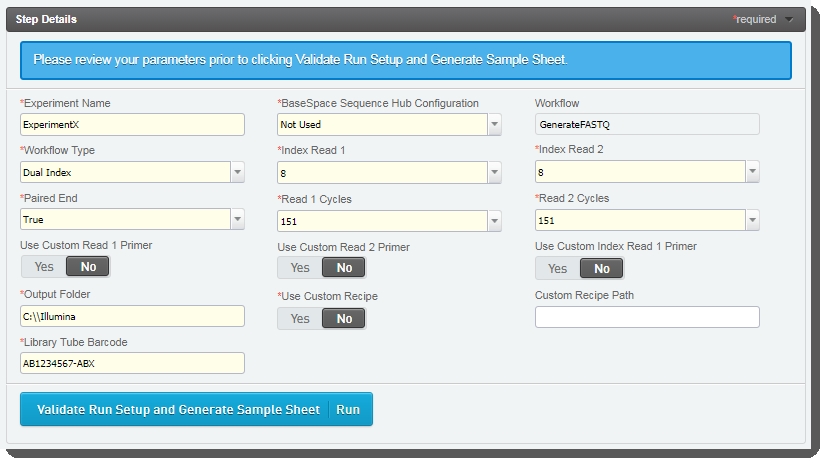
Some of these fields are auto-populated and some must be completed by the user (see the following table for details):
|
Field |
Value |
|
Experiment Name |
User enters the experiment name. Only alphanumeric characters, dashes, and underscores are permitted. Spaces are not permitted. |
|
BaseSpace Sequence Hub Configuration |
User selects from preset options: Not Used, Run Monitoring Only, Run Monitoring and Storage |
|
Workflow |
Automatically populated with preset - GenerateFASTQ |
|
Workflow Type |
User selects from preset options: No Index, Single Index, Dual Index, Custom |
|
Index Read 1 |
User selects from preset options: 0, 6, 8 - or type a value between 0 and 20 |
|
Index Read 2 |
User selects from preset options: 0, 6, 8 - or type a value between 0 and 20 |
|
Paired End |
User selects from preset options: True or False |
|
Read 1 Cycles |
User selects from preset options: 251, 151, 101, 51 - or type a value between 1 and 251 Value of 251 is only supported for SP flow cell type. For all other flow cell types, maximum value is 151. |
|
Read 2 Cycles |
User selects from preset options: 251, 151, 101, 51 - or type a value between 1 and 251 Value of 251 is only supported for SP flow cell type. For all other flow cell types, maximum value is 151. |
|
Use Custom Read 1 Primer |
User selects if applicable. |
|
Use Custom Read 2 Primer |
User selects if applicable. |
|
Use Custom Index Read 1 Primer |
User selects if applicable. |
|
Output Folder |
User enters the network path for sequencing run folder. For example: \\networkshare\run_data |
|
Use Custom Recipe |
User selects if applicable. |
|
Custom Recipe Path |
If Use Custom Recipe selected, user enters the path to the custom recipe file to be used. |
|
Library Tube Barcode |
User scans the library tube barcode. |
| 4. | On the Record Details screen, user clicks Validate Run Setup and Generate Sample Sheet. |
This triggers the automation script, which does the following:
| • | Validates the information provided to set up the run. |
| • | Generates the sample sheet file and attaches it to the placeholder in the Files area of the Record Detailsscreen. |
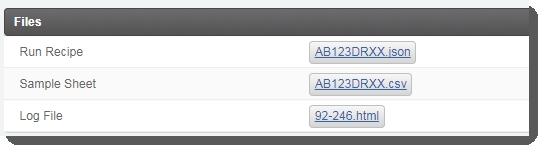
| 5. | User clicks Next Steps. |
| 6. | On the Assign Next Steps screen, the Next Step field for all samples is pre-populated with Mark protocol as complete. User clicks Finish Step. |
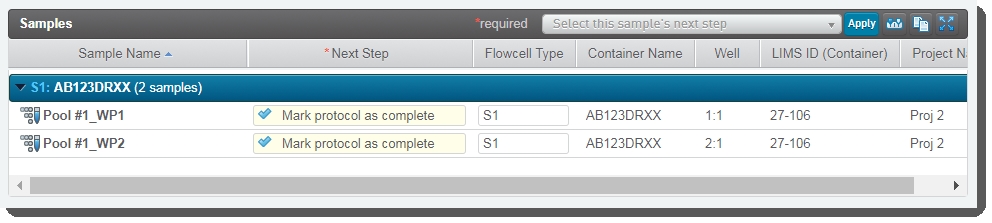
| 7. | In Lab View, the pooled samples are queued for the AUTOMATED - NovaSeq Run (NovaSeq 6000 v3.0) step. |
At this point in the workflow, the user interaction ends. The integration automatically starts the AUTOMATED - NovaSeq Run (NovaSeq 6000 v3.0) step and data from the run is parsed back into the LIMS.
Protocol 4: AUTOMATED - NovaSeq Run (NovaSeq 6000 v3.0)
This final protocol contains one fully-automated step, AUTOMATED- NovaSeq Run (NovaSeq 6000 v3.0), described below.
This is a fully-automated step. The user must not add samples to the Ice Bucket or start the step manually.
Step 1: AUTOMATED - NovaSeq Run (NovaSeq 6000 v3.0)
Step input: Library tube from NovaSeq Standard or flow cell from NovaSeq Xp protocol
Step output: Result file/measurement
The user must not start this step. This is a fully-automated step and the sequencing service will not update samples correctly if they have been manually started.
In this step, pooled samples are sequenced on the NovaSeq instrument and the run metrics are recorded in the LIMS.
The Set Next Steps automation is the only automation configured on this step. By default, this automation is disabled (set to Not Used) and the Sequencer API is used to set the next step for samples.
AUTOMATED - NovaSeq Run (NovaSeq 6000 v3.0) automation configuration - LIMS v4.2.x:
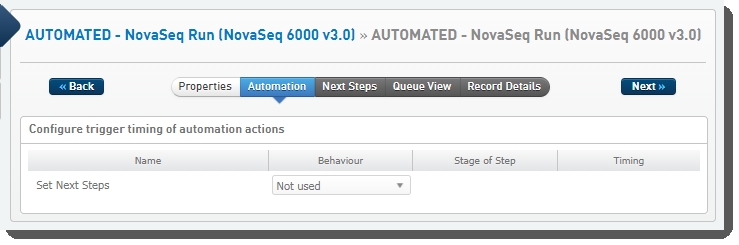
AUTOMATED - NovaSeq Run (NovaSeq 6000 v3.0) automation configuration - LIMS v5.x:

If required, you can override the default next step behavior used by the Sequencer API. To do this:
| 1. | Enable the automation and use it to set custom next step behavior and generate a log file. By default, the automation will set the next step to ADVANCE and the samples complete the protocol. |
| 2. | Configure the automation to be triggered automatically on exit of the Record Details screen. |
| • | If you do not want to override the default next step behavior used by the Sequencer API, you can do one of the following: |
| – | Leave the Set Next Steps automation in place, but disabled at the step level - as configured in the out-of-the-box integration. |
| – | Delete the Set Next Steps automation. |
| • | If you would like to override the default next step behavior used by the Sequencer API, you can do one of the following: |
| – | Enable the automation and use it to set custom next step behavior and generate a log file - as described above. |
| – | Disable/configure auto-complete behavior by setting the values of the run.autoComplete and autoCompleteOnlyAtSuccess properties. See Configuring autoComplete Properties section, below. |
Step UDFs/Custom Fields
The following table shows how the run parameters parsed from the RunParameter.xml file map to step UDFs/custom fields configured on the AUTOMATED - NovaSeq Run (NovaSeq 6000 v3.0) step in the LIMS.
|
Run Parameter |
Step UDF/Custom Field Name |
|
currentCycle |
Current Cycle |
|
currentRead |
Current Read |
|
firmwareVersion |
Firmware Version |
|
FlowCellExpirationDate |
Flow Cell Expiration Date |
|
FlowCellId |
Flow Cell ID |
|
FlowCellLotNumber |
Flow Cell Lot Number |
|
FlowCellMode |
Flow Cell Mode |
|
FlowCellPartNumber |
Flow Cell Part Number |
|
FlowCellSide |
Flow Cell Side |
|
controlSoftwareVersion |
Instrument Control Software Version |
|
instrumentId |
Instrument ID |
|
instrumentType |
Instrument Type |
|
WorkflowType |
Loading Workflow Type |
|
OutputRunFolder |
Output Folder |
|
rtaVersion |
RTA Version |
|
RunId |
Run ID |
|
runStatus |
Run Status |
|
sequencingLog |
Sequencing Log |
Global UDFs/Custom Fields
The following global UDFs/custom fields are used to capture the run metrics in the LIMS.
| • | % Aligned R1 |
| • | % Aligned R2 |
| • | % Bases >=Q30 R1 |
| • | % Bases >=Q30 R2 |
| • | % Error Rate R1 |
| • | % Error Rate R2 |
| • | % Phasing R1 |
| • | % Phasing R2 |
| • | % Prephasing R1 |
| • | % Prephasing R2 |
| • | %PF R1 |
| • | %PF R2 |
| • | Cluster Density (K/mm^2) R1 |
| • | Cluster Density (K/mm^2) R2 |
| • | Intensity Cycle 1 R1 |
| • | Intensity Cycle 1 R2 |
| • | Reads PF (M) R1 |
| • | Reads PF (M) R2 |
| • | Yield PF (Gb) R1 |
| • | Yield PF (Gb) R2 |
Note the following:
| • | Values are aggregated across all lanes. Some values - for example Yield PF (Gb) R1 - are summed, while others are averaged. |
| • | The names listed above are the default UDF/global field names installed with the NovaSeq Integration v3.2.1 configuration provided in the Preconfigured Workflow Package (PCWP) v1.2 or later. |
| • | All UDFs/global fields are configured on the Container entity. |
| • | All field names are configurable via the LIMS Configuration > Custom Fields screen (Global Fields tab). |
| • | If you change the field names in the LIMS, you must also change them through the run.metricUdfNames properties file in the application.yml file for the Sequencer API (see application.yml Properties). All fields are configured to be visible on the AUTOMATED - NovaSeq Run (NovaSeq 6000 v3.0) step's Record Details screen, in the Sample Details table. |
| • | As these UDFs/fields are configured on the Container entity, they are visible on each input. However, they should not be confused with per-lane metrics. |
| • | All run metrics tracked in the LIMS are overall metrics for the run; there are no per-lane metrics provided by NovaSeq at this time. |
How the Sequencer API Integration Works
The sample sheet is generated on the step prior to the run – Dilute and Denature (NovaSeq 6000 v3.0) in the NovaSeq Standard protocol or Load to Flowcell (NovaSeq 6000 v3.0) in the NovaSeq Xp protocol. This is the step where samples are placed on the library tube or flow cell that will be loaded in the NovaSeq instrument.
In the out-of-the-box configuration, the Validate Run Setup and Generate Sample Sheet automation generates one CSV format sample sheet file for use with bcl2fastq v2.20 downstream analysis.
How it Works
The sample sheet is uploaded to the NVCS via the /Illumina/Sequencer/v2/sequencing-run/files endpoint to the Sequencer API. The files endpoint allows for a file to be downloaded from the LIMS using OAuth (instead of Basic Authentication), which is required for NovaSeq.
The run recipe response sets the sample sheet URL to the link to download the file from this endpoint, and sets the sampleSheetRequiresOAuth value to true.
Sample Sheet Contents
The sample sheet content is controlled by the following step UDFs/master step fields that display in the Step Details area of the Record Details screen.
| • | Experiment Name |
| • | Read 1 Cycles |
| • | Read 2 Cycles |
| • | Workflow |
See the Bcl2fastq2 Sample Sheet Generation article for detailed information on the following:
| • | Sample sheet generation script parameters and usage |
| • | Sample sheet data and configuration options |
| • | Enabling unique FASTQ file names per sequencing run |
The steps below outline the sequence of events that occurs when a flow cell is loaded onto the NovaSeq instrument.
The LIMS tracks per-instrument sequencing runs by appending the instrument name to the Run ID field value.
If the InterOp data does not include an InstrumentName value, capturing of run metrics proceeds and a warning is logged. If the InterOp data does include an InstrumentName value, and multiple instruments with the same name exist in the LIMS, capturing of run metrics fails and an error is logged.
User authentication and login to the NovaSeq instrument is achieved via the getSequencerLoginURL endpoint (found at /Illumina/Sequencer/v2/sequencing-run/login). This endpoint returns the URL that the sequencer uses to get an access token.
| 1. | The Sequencer API service provides the NovaSeq Control Software (NVCS) with a URL for the LIMS login screen and the NVCS displays this screen to the user. When the user logs in, this automatically authorizes the NVCS to use their account to access the LIMS. |
| 2. | User enters credentials: |
| • | Username and password are passed to the Clarity LIMS server, to ensure the access is valid. |
| • | By default, the Sequencer API service tries to contact the LIMS at http://localhost:9080/clarity/. This URL is set by the installation script, but can be configured with the clarity.url property in the application.yml file (see application.yml Properties). |
| 3. | User is redirected to the Sequencer login page at /Illumina/Sequencer/v2/sequencer_login. |
| • | If authentication is successful, the screen displays a success message. The LIMS server provides the OAuth token and username by returning them in redirect to /Illumina/Sequencer/v2/sequencer_login/Success. This redirect does not reload the page, but allows the NovaSeq to continue with run setup. |
| • | If authentication fails, the user is left on the login screen and a warning message informs them of the failure. A background redirect to /Illumina/Sequencer/v2/sequencer_login/Error informs the NovaSeq of the failure. |
Respond to Validation/Recipe Request Call from Sequencer
| 1. | The sequencing-run/recipe/novaseq endpoint consumes a POST request with a LibraryContainerI, FlowCellId, and reagents information. |
| • | LibraryContainerId, FlowCellId, and reagents information are provided by the NovaSeq. LibraryContainerId is required. |
| • | LibraryContainerId and FlowCellId should match either the library tube ID or flow cell ID of the corresponding container(s) in the LIMS. |
| • | If there are no containers or multiple containers, an error is generated. |
| 2. | All populated containers whose name matches either the LibraryContainerId or FlowCellId are validated as follows: |
| • | All samples in the container must be queued for a sequencing run step. |
| • | All samples must be queued for only one sequencing run step. |
| • | All samples must be queued for the same sequencing run step. |
You can configure the steps that are considered as 'sequencing run steps' in the novaseq.sequenceStepNames property in the application.yml file. The property is a list and each list entry must be the exact name of a sequencing step, not the name of the underlying master step/process type. See application.yml Properties.
| 3. | Once a single container has passed validation, the run setup information (run recipe request) is returned to the NVCS via the API and the run recipe is generated. |
Note the following:
| • | The run recipe information is read from fields configured on the parent process/master step of the samples in the container. The workflow must include recipe definition in that step. |
| • | By default, the parent process/master step is the Dilute and Denature (NovaSeq 6000 v3.0) or Load to Flowcell (NovaSeq 6000 v3.0) step, depending on workflow. |
| • | The default field values match the names of the fields in the NovaSeq v3.2.1 configuration included in PCWP. The field names are configurable through the recipe.udfNames.* properties. Each of these properties corresponds to a entry in the recipe and the value of a property defines the name of the field that is used to populate that entry in the recipe. |
| • | It is assumed that this parent process/master step removes any illegal characters from the Experiment Name field. |
| • | The LIMS download link to the sample sheet created on the run setup step is used for the SampleSheet value in the recipe. The name of the output of the sample sheet on the setup step can be configured through the recipe.sampleSheet.outputName property (Sample Sheet by default) in the application.yml (see application.yml Properties). If no matching output is found, or no file was attached to the output, then the value of the recipe.sampleSheet.notAvailableValue property (Not Available by default) is used in the recipe. |
| 4. | After the recipe is generated, it is returned as a *.json file in the response, in the form defined by the Swagger definition for the API. |
The contents of the run recipe are controlled by the sample sheet contents and the step UDFs/master step fields, configured on the Dilute and Denature (NovaSeq 6000 v3.0) and Load to Flowcell (NovaSeq 6000 v3.0) steps:
Example of run recipe content is as follows:
{
"run_name": "NVCS16027_SPrime_2x250",
"run_mode": "S4",
"workflow_type": "DualIndex",
"librarytube_id": "NV0025867-LIB",
"flowcell_id": "",
"sample_loading_type": "NovaSeqStandard",
"rehyb": false,
"paired_end": true,
"read1": 151,
"read2": 151,
"index_read1": 8,
"index_read2": 8,
"output_folder": "\\\\network_path\\run_data",
"samplesheet": "<path to sample sheet on LIMS server>",
"require_samplesheet_authentication": true,
"usecustomrecipe": false,
"customRecipe": null,
"use_basespace": false,
"basespace_mode": null,
"use_custom_read1_primer": false,
"use_custom_read2_primer": false,
"use_custom_index_read1_primer": false
}
Respond to Sequencing Started Call from Sequencer
| 1. | Matching container for the run (library tube or flow cell) is found, and contents are validated to be queued for the correct step. |
An error is thrown and the step is not started if no matching container is found, or if the samples in the container are incorrectly queued. This validation is also done during the run recipe request immediately prior, so this error case should not be encountered.
| 2. | Reagent kits that are configured for the Sequencer API are checked against the configuration for the step to be run: |
| • | If a kit is configured for the API but does not exist in LIMS, it is created. |
| • | If a kit exists, but is not enabled on the step to be run, the step configuration is updated to enable it |
| – | If a kit that is to be enabled is Archived, it is set to Active. |
| – | If a kit that is to be enabled is in Pending status, it is set to Active. |
| • | If any kits enabled on the step are not configured for the Sequencer API, a warning is logged and the step is unable to be completed by the API without user intervention. |
| • | If any kits provided in the recipe request are not configured for the Sequencer API, they are ignored and a warning is logged during lot tracking. |
| 3. | The number of replicates for the step is determined by the type of flow cell being used in the run. |
| • | For Standard mode, this is based on the number of lanes in the flow cell (this is stored as part of the Sequencer API configuration). |
| • | For XP mode, this is always one replicate, since there is already one input tracked for each lane. |
| 4. | The automated sequencing step is started. |
| 5. | Reagent lots are attached to the step. |
| • | The Flow Cell and Library Tube reagents in the request are ignored - information from these is tracked in the LIMS as containers and UDFs. |
| • | Any other reagents in the request are handled as follows:If the reagent is not configured to be tracked, a warning is logged and the reagent is ignored. |
| – | Existing reagent lots are looked up by kit name (determined by Sequencer API configuration), name (the serial number in the request), and lot number (the lot number in the request). |
| – | If one or more matching reagent lot exists in the LIMS, the newest one is used: |
| – | If the expiry date does not match the expiry in the request, or if the status is not set to Active, the lot is updated to have the correct expiry date and an Active status. |
| – | Otherwise a new lot is created with: |
| – | Reagent Kit: Matching kit as determined from Sequencer API configuration and step 2 above. |
| – | Name: Serial number in request |
| – | Lot Number: Lot number in request |
| – | Expiry: Expiry date in request |
| – | Status: Active |
| • | If any reagents were expected to be in the request but were not, these reagents are logged. This prevents the step from being completed without user intervention. |
| 6. | Step UDFs/master step fields are updated with the run information parsed from the request. See Respond to Record InterOps Metrics Call from Sequencer, below. |
| 7. | Any important events during this request are logged into the Sequencer API log file as well as in the Sequencing Log multiline UDF/field on the step (assuming the step was started successfully). See Respond to Record InterOps Metrics Call from Sequencer, below. |
Respond to Record InterOps Metrics Call from Sequencer
| 1. | When the metrics endpoint receives a POST, it: |
| • | Looks for a matching library tube or flow cell in the LIMS. |
| • | Find finds the in-progress sequencing step for that container. |
| 2. | When the step is found, the run metrics in the request are written to container UDFs/global fields in the LIMS. |
Respond to Sequencing Completed Call from Sequencer
If the sequencing run was successful, after the metrics have been recorded, the API call for Sequencing Completed is received. Information is matched by Sequencing Container and the following occurs in the LIMS:
| 1. | The Current Cycle, Current Read, and Status UDFs/master step fields are updated. |
| 2. | The multiline Sequencing Log UDF/master step field is updated. |
| 3. | A success message is recorded in the log file, at the info level: |
Sequencing run with Run ID {runId} of Step '{step.name}' ({step.limsid}) completed successfully.
| 4. | The LIMS waits for any automations that are triggered automatically as it advances the step. |
| 5. | By default, the LIMS assigns the next steps on the outputs to move them on to the first configured next step - or, if there are no next steps, to complete the protocol. |
Respond to Sequencing Errored Call from Sequencer
If the sequencing run was unsuccessful, the API call for Sequencing Error is received. Information is matched by Sequencing Container and the following occurs in the LIMS:
| 1. | The Current Cycle, Current Read, and Status UDFs/master step fields are updated. |
| 2. | The multiline Sequencing Log UDF/master step field is updated. |
| 3. | A failure message is recorded in the log file, at the error level: |
Step '${Step.name}' (${Step.limsid}) failed with Run ID ${runId}.
| 4. | The step is left at the Record Details screen. |
Respond to Sequencing RunEndedByUser Call from Sequencer
If the user ends the sequencing run, the API call for Sequencing RunEndedByUser is received. Information is matched by Sequencing Container and the following occurs in the LIMS:
| 1. | The Current Cycle, Current Read, and Status UDFs/master step fields are updated. |
| 2. | The multiline Sequencing Log UDF/master step field is updated. |
| 3. | The following message is recorded in the log file, at the warning level: |
"Sequencing run with Run ID ${status.runInfo.runId} of Step '${step.configuration.name}' (${step.limsid}) aborted by user ${status.runInfo.userName}."
| 4. | The step is left at the Record Details screen. |
Components Installed with this Integration
The following sections describe the various components - files, properties, reagent categories / label groups, reagent kits, and containers - that are installed by default as part of this integration. Information on installed workflows, protocols, steps, and automation points is provided in the Workflows, protocols, and steps section, above.
Illumina NovaSeq Integration v3.2.1 is distributed as an RPM package - BaseSpaceLIMS-sequencer-api. This RPM package must be installed on the LIMS server.
The BaseSpaceLIMS-sequencer-api RPM installs the following:
| • | Sequencer API WAR file |
| • | Configuration file - application.yml |
| • | Two configuration scripts - configure_sequencer_api_proxy.sh and configure_sequencer_api_application.sh |
The NovaSeq v3 configuration is delivered through the Preconfigured Workflows Package (PCWP) v1.2, which requires installation of the NGS Extensions Package v5.18 or later.
| • | If the NGS Extensions Package is not already installed, or if a version earlier than v5.18 is installed, the latest version is installed by default with the NovaSeq integration. See Release Notes for details. |
| • | If NGS Extensions Package v5.18 or later is already installed, upgrade is not forced or required. |
The RPM package installs the components listed in the following table:
|
Files Installed |
Location and Description |
|
Illumina#Sequencer#v2.war |
/opt/gls/clarity/tomcat/current/webapps War file for NovaSeq v3.2.1 API |
|
configure_sequencer_api_application.sh |
/opt/gls/clarity/config/ Script that configures the Sequencer API application through its external application.yml file |
|
configure_sequencer_api_proxy.sh |
/opt/gls/clarity/config/ Script that configures the proxy to allow communication with the Sequencer API (NovaSeq v3.2.1 API) application |
|
sequencer-api.conf |
/etc/httpd/clarity/ proxy setting to communicate with the LIMS |
The application.yml file is located at opt/gls/clarity/extensions/sequencing-api/.
| • | All properties are configured automatically during installation, but you can also configure them by editing the application.yml file and then restarting Tomcat. |
| • | All custom field names, both for recipe and run information, are represented by properties in the file. |
| • | For all custom fields, the names (values in this file) can be changed. However, the property names cannot be changed, removed, or added to. For example, you can change Loading Workflow Type but you cannot change recipe.udfNames.sampleLoadingType. |
The properties it contains are listed in the following table:
|
Property |
Description |
|||||||||
|
spring.profiles.active |
Tells the application that it is running deployed in Tomcat. Default value: "tomcat" DO NOT CHANGE |
|||||||||
|
clarity.url |
Base URL that the Sequencer API service uses to contact the LIMS. The installation script (configure_sequencer_api_application) prompts for this. |
|||||||||
|
clarity.username |
Username to be used when communicating with Clarity LIMS |
|||||||||
|
clarity.password |
Encrypted password to be used when communicating with Clarity LIMS |
|||||||||
|
security.signing-key |
Private key that should be used when signing/validating OAuth tokens. Changing this will invalidate any issued tokens. |
|||||||||
|
security.token-expiry |
Specifies (in hours) the expiry period for login tokens issued by the Sequencer API. Default value: 88 |
|||||||||
|
novaseq.sequenceStepNames |
List of NovaSeq sequencing run step names for which the integration may find samples queued.
Default value: - "AUTOMATED - NovaSeq Run (NovaSeq 6000 v3.0)" |
|||||||||
|
novaseq.flowcells |
List of supported NovaSeq flow cell types.
Default value: - type: "SP"lanes: 2- type: "S1"lanes: 2- type: "S2"lanes: 2- type: "S4"lanes: 4 |
|||||||||
|
novaseq.reagents |
List of supported reagent kits to be tracked on sequencing steps. - lotName: The name of the lot as it appears in the run started request. - kitName: The name of the corresponding reagent kit in LIMS that the lot should be tracked as.
Default value: - lotName: "Buffer"kitName: "Buffer Cartridge"- lotName: "Cluster"kitName: "Cluster Cartridge"- lotName: "SBS"kitName: "SBS Cartridge" |
|||||||||
|
recipe.udfNames |
Configures the names of the UDFs/fields that will be used in the run recipe (see Run Recipe Contents for more information).
|
|||||||||
|
recipe.sampleSheet.outputName recipe.sampleSheet.notAvailableValue |
Configures the name of the sample sheet file placeholder on the step where run recipe information is populated, and the property value to use when the file is not found. |
|||||||||
|
run.stepUdfNames |
Configures the step UDF/master step field names that will be used when recording run results (see Step UDFs/Custom Fields for more information).
|
|||||||||
|
run.metricUdfNames |
Configures the result file UDFs/measurement global fields names that will be used when recording run results. These are the fields that capture the run data (see Global UDFs/Custom Fields for more information). |
|||||||||
|
run.autoComplete |
Determines whether the sequencing run step should be completed automatically after parsing the run metrics on a run (see also run.autoCompleteOnlyAtSuccess). Default value: true See Configuring autoComplete Properties, below. |
|||||||||
|
run.autoCompleteOnlyAtSuccess |
Determines whether the sequencing run step should be:
- or -
Default value: true See Configuring autoComplete Properties, below. |
Reagent Categories/Label Groups
| • | TruSeq HT Adapters v2 (D7-D5) |
| • | Buffer Cartridge |
| • | Cluster Cartridge |
| • | DPX1 |
| • | DPX2 |
| • | DPX3 |
| • | NaOH |
| • | Resuspension Buffer (RSB) |
| • | SBS Cartridge |
| • | Tris HCl |
| • | Library Tube |
| • | SP |
| • | S1 |
| • | S2 |
| • | S4 |
This integration supports:
| • | Library tube with barcode provided in the format [A-Za-z]{2}[0-9]{7}-[A-Za-z]{3} |
Example: AB1234567-XYZ
| • | SP, S1, S2, or S4 flow cell, with barcode provided in one of the following formats: |
| – | [A-Za-z0-9]{5}DRXX |
| – | [A-Za-z0-9]{5}DMXX |
| – | [A-Za-z0-9]{5}DSXX |
Example: AB123DRXX
For details on configuring NVCS for integration with BaseSpace Clarity LIMS, contact the Clarity LIMS Support team.
The requirements for the routing script functionality are as follows:
| • | On the steps that use the routing script - Define Run Format (NovaSeq 6000 v3.0) and Dilute and Denature (NovaSeq 6000 v3.0) - the Next Step for all samples must be set to Remove from workflow. This value is set by a script, and the user running the step must not change this value in the Assign Next Steps screen. |
| • | LIMS v5.x: In the protocol configuration screen, the following settings are required and must not be changed: |
In the Next Steps section, for the last step in the protocol, the method of assigning the next step must be set to Automatic.
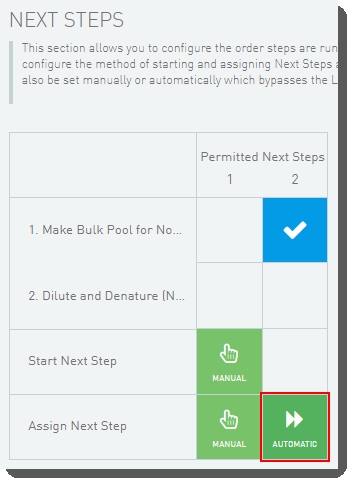
| • | LIMS v4.2.x: In the protocol configuration area, for all protocol steps on which the Routing Script automation is configured, the following setting is required and must not be changed: |
On the Next Steps configuration tab, the Next Steps are set by a script toggle switch must be set to Yes.
Configuring autoComplete Properties
As of NovaSeq Integration v3.2.1, the application.yml configuration file contains two additional properties:
| • | run.autoComplete |
| • | run.autoCompleteOnlyAtSuccess |
These properties determine the conditions under which the last step of the NovaSeq 6000 workflow - AUTOMATED - NovaSeq Run (NovaSeq 6000 v3.0) - is automatically completed:
| • | Only when the run status is "RunCompletedSuccessfully" (default behavior) |
-or -
| • | Regardless of the run status |
By default, both properties are set to 'true' and the step only completes if the run is successful. If the run fails or is aborted by the user, the user must manually complete the step in the LIMS.
If you would like the step to autocomplete regardless of the result of the sequencing run, you must change the autoCompleteOnlyAtSuccess property value to 'false'.
Enabling/Disabling autoComplete Properties
| 1. | Open the application.yml file located at /opt/gls/clarity/extensions/sequencer-api/application.yml: |
vi /opt/gls/clarity/extensions/sequencer-api/application.yml
| 2. | Edit the value of the autoCompleteOnlyAtSuccessproperty as required. |
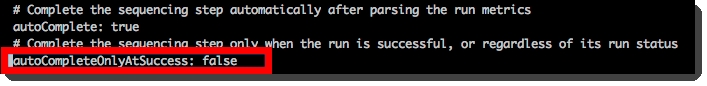
| 3. | Save the file. |
The following table shows how the combined value of the run.autoComplete and run.autoCompleteOnlyAtSuccess properties affects the auto-complete behavior of the sequencing step.
|
run.autoComplete property value |
run.autoCompleteOnlyAtSuccess property value |
Outcome |
||||||
|
true (default) |
true (default) |
|
||||||
|
true |
false |
|
||||||
|
false |
true |
|
||||||
|
false |
false |
| • | The workflow configuration contains several validation checks. To ensure that the calculations work properly, it is important that you do not disable any of this validation logic. The validation checks determine: |
| – | Which samples, and how many, can enter each step together. |
| – | Which samples, and how many, can be pooled together. |
| • | The library tube ID must be unique. There should not be multiple library tube containers in the system with the same name. |
| • | Reagent labels (indexes) must be unique. |
| • | Only controls are permitted as unindexed samples; all other unindexed samples and pools are not permitted. |
| • | For sample sheet generation constraints, refer to the Bcl2fastq2 Sample Sheet Generation article. |
| • | Users must not manually start the AUTOMATED - NovaSeq Run (NovaSeq 6000 v3.0) step. This is a fully-automated step and the sequencing service will not update samples correctly if they have been manually started. |
| • | For the automated run to start successfully, the user must have clicked the Validate Run Setup and Generate Sample Sheet button. |
| • | NovaSeq 6000 Sequencing System Guide |
